Figure 6. Role of PI3K in NF-κB-dependent MAT2A expression in TAMR-MCF-7 cell.
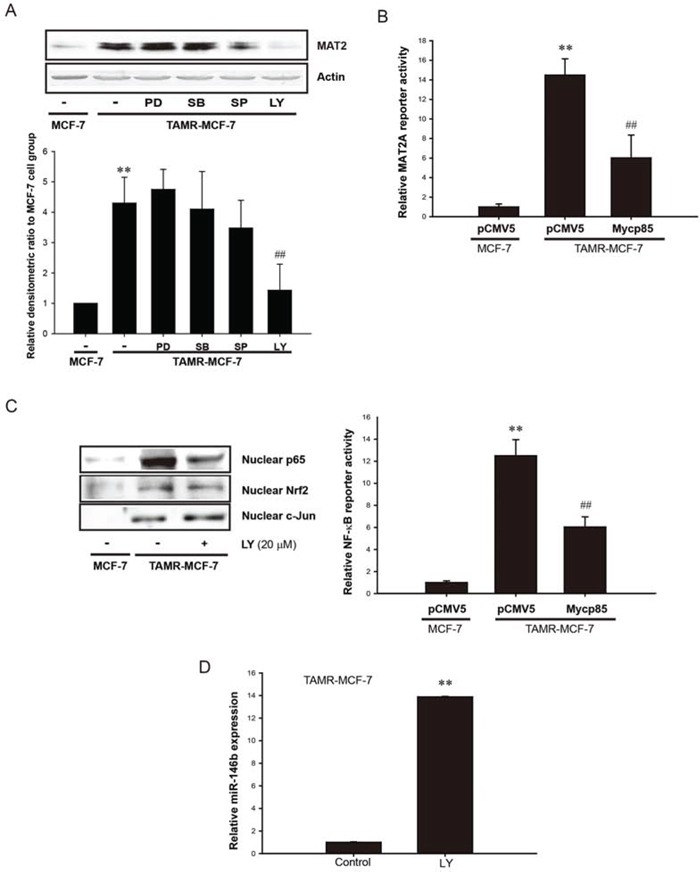
A. Effect of MAPK inhibitors and PI3K inhibitor on MAT2A expression. Upper; MAT2A protein expression was completely suppressed by 24 h incubation of LY294002 (LY, 20 μM), a PI3K inhibitor in TAMR-MCF-7 cells. PD98059 (PD, 20 μM, ERK inhibitor), SB203580 (SB, 10 μM, p38 kinase inhibitor) and SP600125 (SP, 10 μM, JNK inhibitor) were also used. Lower; Densitometry data. Data represent mean ± SD with 3 different samples (significant versus control MCF-7 cells, **P < 0.01; significant versus TAMR-MCF-7 cells, ##P < 0.01). B. Effect of Mycp85 (dominant negative mutant form of PI3K) overexpression on MAT2A gene transcription. TAMR-MCF-7 cells were co-transfected with MAT2A-luc reporter plasmid (1 μg/well) phRL-SV plasmids (1 ng/well) and pCMV5 or Mycp85 overexpression plasmid (0.5 μg/well) for 18 h in serum free condition. Luciferase reporter activity was determined described as figure legend of Figure 1B. Data represent mean ± SD with 3 different samples (significant versus pCMV5-transfected MCF-7 cells, **P < 0.01; significant versus pCMV5-transfected TAMR-MCF-7 cells, ##P < 0.01). C. Effect of PI3K inhibition on NF-κB activity. Left; nuclear p65, nuclear Nrf2, and nuclear c-Jun detection. TAMR-MCF-7 cells were treated with LY294002 (LY, 20 μM) for 24 h and nuclear levels of p65, Nrf2, and c-Jun were detected by immunoblotting. Right; Effect of Mycp85 overexpression on NF-κB minimal reporter activity. TAMR-MCF-7 cells were co-transfected with pNF-κB-Luc reporter plasmid (1 μg/well) and pCMV5 or Mycp85 overexpression plasmid (0.5 μg/well). Then, pNF-κB-Luc reporter activity was determined 18 h after transfection in serum-free condition. Data represent mean ± SD with 3 different samples (significant versus pCMV5-transfected MCF-7 cells, **P < 0.01; significant versus pCMV5-transfected TAMR-MCF-7 cells, ##P < 0.01). D. Effect of LY294002 (PI3K inhibitor, LY, 20 μM) on miR-146b expression in TAMR-MCF-7 cells. TAMR-MCF-7 cells was exposed to LY294002 for 24 h and then miR-146b level was determined by using miScript PCR. Samples were normalized to small nRNA U6. Data represent mean ± SD with 3 different samples (significant versus vehicle-treated TAMR-MCF-7 cells, **P < 0.01).
