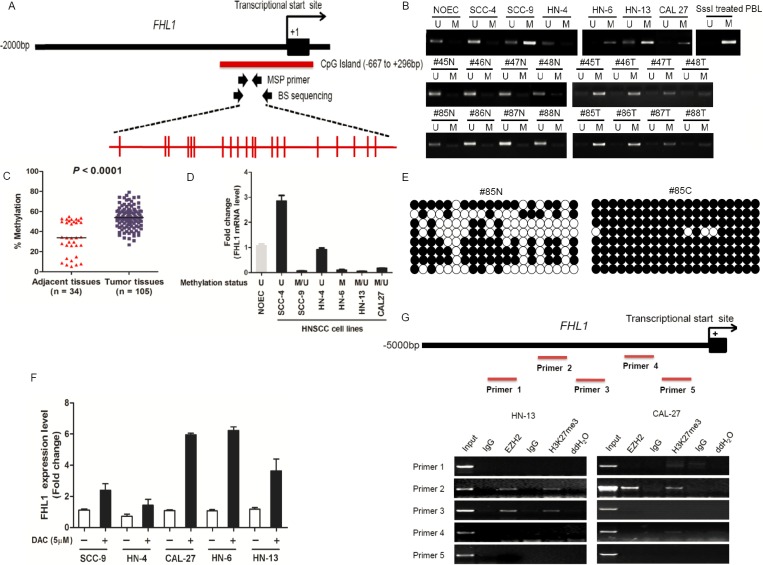
Figure 5. DNA and EZH2-mediated histone hypermethylation together contribute the silencing of FHL1 in HNSCC.
(A) Schematic representations of the location of CpG island within the promoter of FHL1 and of the primers designed against the promoter region for Methylation-specific polymerase chain reaction (MSP-PCR) and bisulfite-treated DNA sequencing (BS). Red ticks indicated CpG site in CpG island region. (B) Methylation status of FHL1 was investigated in normal oral epithelial cell (NOEC), HNSCC cell lines and eight representative paired HNSCCs by MSP-PCR. (C) Methylation status of FHL1 was further revealed in paired adjacent tissues (n = 34) and HNSCCs (n = 105) by MSP-PCR. (D) The negative association between FHL1 mRNA expression and FHL1 methylation status was illustrated. (E) Methylation status of FHL1 was determined in a representative paired adjacent tissue and HNSCC sample by BS. (F) Expression restoration of FHL1 was also observed in five HNSCC cells by real-time PCR after the 5-Aza-dC induction. (G) Schematic representations of the location of putative binding region within the promoter of FHL1 and of the five primers designed against the promoter region for Chromatin Immunoprecipitation (ChIP)-PCR, and notably occupancy of EZH2 and H3k27me3 were found in both HN-13 and CAL-27 cells.