Figure 7. Abnormal expression of miR-410 is not regulated by methylation or acetylation in NSCLC cells.
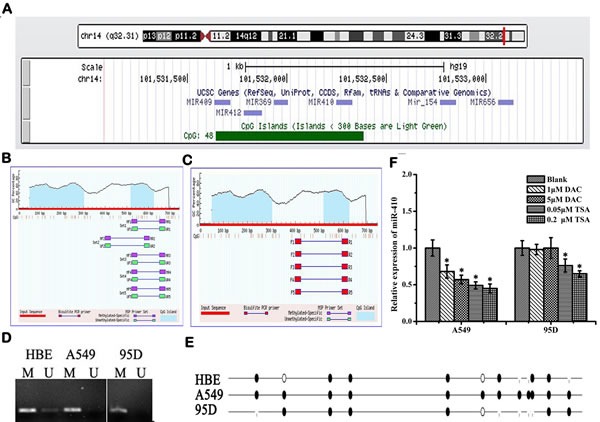
A. CpG island located in the gene locus of miR-410 according to UCSC database (http://genome.ucsc.edu/cgi-bin/hgGateway). Blue areas: predicted CpG islands (Locus: chr14: 101531644-101532384, GC content 67.2%, Obs/Exp value 0.8). (B and C) Primers were designed to cover the CpG islands upward miR-410 for methylation analysis. Primer sets for MSP B. and BSP C. according to MethPrimer (http://www.urogene.org/methprimer/index1.html). D. MSP results detected by 2% agarose gel electrophoresis in A549, 95D and HBE cells. U, unmethylated; M, methylated. HBE, human bronchial epithelial cells. The result indicated that miR-410 was partly methylated in normal HBE and 95D cells, but almost totally methylated in A549 cells. E. Bisulfite sequencing analysis of miR-410 in A549, 95D and HBE cells. Black dot, methylated; White dot, unmethylated. The result of bisulfite sequencing (BSP) showed that all the 12 CpGs sites sequenced were completely methylated in A549 cells, and 6 of 12 CpGs sites (50%) were methylated in 95D cells, and 7 of 12 CpGs sites (58.3%) were methylated in HBE cells, in consistent with the result of MSP. F. Expression levels of miR-410 were detected by qRT-PCR in A549 and 95D cells after treatment with DNA methylation inhibitor 5′-aza-2′-deoxycytidine (DAC) and/or histone deacetylase (HDAC) inhibitor trichostatin A (TSA). Data are expressed as means ± SD of triplicate assays. *, p < 0.05; **, p < 0.01, significant difference vs untreated group.
