Abstract
Accumulating data indicates that chromosome instability (CIN) common to cancer cells can be used as a target for cancer therapy. At present the rate of chromosome mis-segregation is quantified by laborious techniques such as coupling clonal cell analysis with karyotyping or fluorescence in situ hybridization (FISH). Recently, a novel assay was developed based on the loss of a non-essential human artificial chromosome (HAC) carrying a constitutively expressed EGFP transgene (“loss of signal” assay). Using this system, anticancer drugs can be easily ranked on by their effect on HAC loss. However, it is problematic to covert this “loss of signal” assay into a high-throughput screen to identify drugs and mutations that increase CIN levels. To address this point, we re-designed the HAC-based assay. In this new system, the HAC carries a constitutively expressed shRNA against the EGFP transgene integrated into human genome. Thus, cells that inherit the HAC display no green fluorescence, while cells lacking the HAC do. We verified the accuracy of this “gain of signal” assay by measuring the level of CIN induced by known antimitotic drugs and added to the list of previously ranked CIN inducing compounds, two newly characterized inhibitors of the centromere-associated protein CENP-E, PF-2771 and GSK923295 that exhibit the highest effect on chromosome instability measured to date. The “gain of signal” assay was also sensitive enough to detect increase of CIN after siRNA depletion of known genes controlling mitotic progression through distinct mechanisms. Hence this assay can be utilized in future experiments to uncover novel human CIN genes, which will provide novel insight into the pathogenesis of cancer. Also described is the possible conversion of this new assay into a high-throughput screen using a fluorescence microplate reader to characterize chemical libraries and identify new conditions that modulate CIN level.
Keywords: chromosome instability, CIN, human artificial chromosome, HAC, anticancer drugs
INTRODUCTION
Abnormal chromosome number (aneuploidy) is a known feature of most solid tumors and is often accompanied by an elevated rate of chromosome mis-segregation termed chromosome instability (CIN) [1]. The gain or loss of entire chromosomes leads to large-scale changes in gene copy number and expression levels. The molecular mechanisms underlying CIN include defects in chromosome cohesion, mitotic checkpoint function, kinetochore-microtubule attachment dynamics, cell cycle regulation as well as defects in DNA replication. Mutations in CIN genes are thought to be an early event in tumor development, predisposing cells to the accumulation of genetic changes leading to progression to a cancerous state [2-4]. Notably, a significant fraction of human CIN genes remains unidentified and in part this is due to the lack of a simple assay to detect CIN in vertebrate cells, similar to that developed for yeast cells [5]. Thus, there is an important need to develop improved assays for measuring chromosome transmission fidelity in human cells.
While CIN can drive cancer genome evolution and tumor progression, recent findings point to the existence of a threshold level beyond which CIN becomes a barrier to tumor growth. Therefore excessive CIN can be exploited therapeutically [6-11] and evaluation of CIN as an approach to cancer therapy is an attractive strategy. However, drugs known to increase CIN beyond the therapeutic threshold are currently few in number. Hence, a screen of established anticancer drugs as well as novel drugs to rank their CIN potency is warranted.
Typically, rates of chromosome mis-segregation have been quantified by laborious techniques such as coupling clonal cell analysis with karyotyping or fluorescence in situ hybridization (FISH) [12-14] In our recent work, we developed a quantitative assay for measuring CIN [15] that is based on the use of a non-essential human artificial chromosome (HAC) with a functional kinetochore [16-20]. Specifically we used a HAC constructed for gene delivery that contains a single gene-loading site [21-23]. To adapt this HAC for CIN studies, a constitutively expressed EGFP transgene was inserted into it [15]. Cells that inherit the HAC display green fluorescence, while cells lacking the HAC do not (“loss of signal” assay). This allows the measurement of HAC loss rate by routine flow cytometry (details of this assay are shown in Supplementary Figure S1).
There are several advantages of the HAC-based assay compared to karyotype analysis or micronucleus tests that are commonly used to study CIN and its induction by environmental agents. First, the HAC-based assay is significantly faster and less labor intensive. Second, the flow cytometer can readily analyze tens of thousands of cells compared to the hundred or so cells the latter two methods can analyze. Thus, the measurements are more precise. Finally, while the HAC contains a functional centromere/kinetochore and is efficiently transferred at mitosis, its relatively small size (∼1 Mb) [24] causes a frequency of spontaneous HAC loss roughly 10-fold higher than that of native chromosomes [15, 16], making the HAC a sensitized model for measuring CIN. Together, these features of the HAC allow detection of small differences between frequencies of chromosome loss induced by different compounds. This is important because accurate assessment of CIN is crucial to select drugs with the highest effect on chromosome transmission. In our recent study, the EGFP-HAC-based CIN assay was applied for analysis of 62 anticancer drugs corresponding to six groups of compounds with different mechanisms of action [25]. Within each group, drugs could be ranked with regards to their effect on the rate of HAC loss [25].
While EGFP-HAC offers sensitive, precise and simple means to measure CIN, development of a fast high-throughput screening method, based on the detection of fluorescence signal loss is problematic. This is mostly due to the difficulty to detect single cells with a decreased EGFP signal among the large number of the EGFP-positive cells during the first days after drug treatment. Herein, we developed a complementary HAC-based system, based on the gain of the EGFP fluorescence signal after HAC loss. In this system, the HAC carries a constitutively expressed shRNA against an EGFP transgene integrated elsewhere into a natural human chromosome. Thus, cells that inherit the HAC display no green fluorescence, while cells lacking the HAC do. The high precision of this new assay was verified by measuring HAC loss after cell treatment by several anticancer drugs corresponding to three groups of compounds with different mechanisms of CIN induction. In addition, both HAC-based assays have been proven to be applicable for screening human genes to identify phenotypes associated with CIN and thus to identify CIN genes themselves.
RESULTS
Experimental design for quantitative CIN measurement by “gain of signal” assay
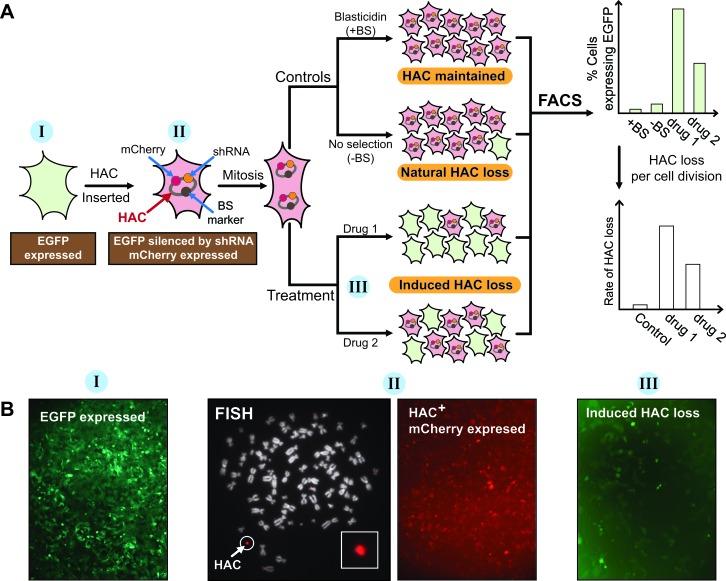
Figure 1A shows the general scheme of the “gain of signal” assay developed for measuring chromosome instability (CIN) using a genetically marked human artificial chromosome (HAC). The EGFP (enhanced green fluorescence protein) transgene is integrated into a random chromosomal site of human HT1080 cells and is expressed, making the cells green (I) (Figure 1A and 1B). The HAC developed in hamster host CHO cells carries the mCherryFP (mCherry fluorescence protein) transgene and a constitutively expressed short hairpin RNA (shRNA) against EGFP. After transfer of the mCherry-shRNA-HAC into HT1080-EGFP-expressing cells, the HAC is maintained as a non-essential 47th chromosome (II) due to the presence of its functional kinetochore and the cells maintaining the HAC become red (II) (Figure 1A and 1B). Normally, after growing in non-selective medium (−BS) (i.e. in the absence of selection for HAC), the majority of cells still retain the HAC (natural HAC loss). However, if proper chromosome transmission is impaired, the fraction of cells that lost the HAC is significantly increased. Thus, while the control untreated cells do not display green fluorescence the cells that have lost HAC after drug treatment (induced HAC loss) should exhibit green fluorescence (III) that can be detected by fluorescence microscopy or flow cytometry (Figure 1A and 1B). In principle, this new assay may provide a convenient and sensitive way to monitor CIN in human cells.
Figure 1. “Gain of signal” HAC-based assay to measure CIN level in cancer cells.
A. A general scheme of assay to measure chromosome instability (CIN), based on the HAC expressing a small hairpin RNA (shRNA) against the EGFP transgene integrated into a random chromosomal site of genome of the HT1080 human cell line. When EGFP is expressed it makes the cell green. Cells containing the mCherry-shRNA-HAC display a red signal. When cells loose the HAC, they become green again. Thus, it is expected that the control population of untreated cells show uniform red fluorescence while cell population that have lost HAC after drug treatment should show green fluorescence. The percentage of cells with EGFP expressed can be measured by FACS. Thus, the compounds, which increase HAC loss and, therefore, increase spontaneous chromosome mis-segregation rates, may be identified. B. Fluorescence images of human HT1080 cells expressing the EGFP transgene (exhibiting a green fluorescent signal) (I); the cells after transfer into them the mCherry-shRNA-HAC (exhibiting a red fluorescent signal) (II) and the same cells that have lost the HAC after drug treatment (exhibiting a green fluorescent signal) (III). FISH analysis of the HAC-containing HT1080 clone is also shown. The HAC was visualized using the BAC32-2-mer(tetO) DNA probe (red) (see for details MATERIALS AND METHODS).
Construction of a cell line for CIN phenotype screening
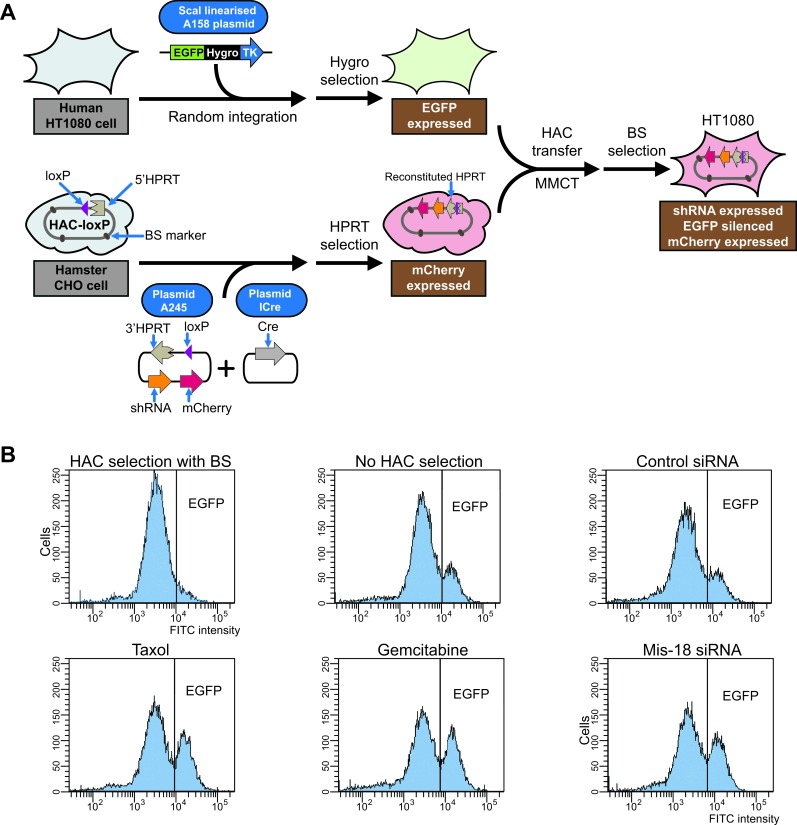
To adapt the alphoidtetO-HAC [16] for CIN studies, the tDNA-shEGFP-mCherry plasmid (A245) (Supplementary Figure S2) was inserted into a single loxP loading site of the HAC [22] in HPRT-deficient Chinese hamster ovary (CHO) cells (Figure 2A). In the A245 construct, the mCherry-shRNA cassette is flanked by tDNA insulator sequences to protect the transgene from epigenetic silencing [26, 27]. Targeting of the cassette into the loxP site was accompanied by reconstitution of the HPRT gene, allowing cell selection on HAT medium. PCR analysis with specific primers (Supplementary Table S1) confirmed that the HPRT gene was indeed reconstituted in five randomly chosen drug-resistant clones. Based on fluorescence microscopy, all clones expressed mCherry. FISH analysis of CHO metaphase spreads revealed the HAC in an autonomous form in three analyzed clones (Supplementary Figure S3). One clone was chosen for further experiments.
Figure 2. Development of “gain of signal” assay and its actual application for drugs screening.
A. General steps of development of “gain of signal” HAC-based assay. First, the EGFP transgene (plasmid A158) was integrated into a random chromosome site in human HT1080 cells, providing a stable expression of the EGFP protein (cells are green). Separately, the tDNA-shEGFP-mCherry plasmid (A245) was inserted into a unique gene-loading loxP site of alphoidtetO-HAC propagated in hamster CHO cells (cells are red). Then alphoidtetO-HAC carrying a constitutively expressed shRNA against EGFP was transferred into human HT1080 cells with the integrated EGFP transgene. After HAC transfer and shRNA expressed, the EGFP transgene is silenced and the cells become red. B. Flow cytometry histograms illustrating EGFP fluorescence before and after drug treatment. The x-axis represents the intensity of the fluorescence, the y-axis the number of cells. The results of triplicate experiments are shown.
The mCherry-shRNA-HAC was transferred from hamster CHO to the human HT1080 cells in which the EGFP transgene (plasmid A158) (Supplementary Figure S4) was integrated into a random chromosome site providing stable expression of the EGFP protein (clone HT1080-JH1) (Figure 2A). The HAC transfer was performed via microcell-mediated chromosome transfer (MMCT). Recipient cells were selected using the BS resistance gene on the alphoidtetO-HAC [16]. Three BS-resistant clones were isolated from one MMCT experiment and all three exhibited barely detectable expression of EGFP, indicating transgene silencing induced by the shRNA. FISH analysis showed that the HAC was maintained autonomously without detectable integration into the host genome in two out of three selected clones (Supplementary Figure S5A). One clone containing the mCherry-shRNA-HAC (clone HT1080-shGFP-4) was chosen for further analysis. In this clone, EGFP silencing was stable for at least six months under selective conditions (i.e. under selection for BS). Rare events of spontaneous HAC loss were accompanied by visual expression of EGFP. Based on these results, we conclude that the mCherry-shRNA is stably expressed from the HAC and that these cells may be exploited as a sensitized system for analyzing chromosome stability.
Quantitative profiling of CIN induced through treatment by anticancer agents
We next investigated whether the mCherry-shRNA-HAC-based assay could be used to quantify the effects of compounds that cause chromosome loss or mis-segregation. For this purpose, we first chose seven anticancer drugs that have been previously ranked on their effects on HAC loss in HT1080 cells using the original “loss of signal” EGFP-HAC-based system (Supplementary Figure S1) [15, 25]. The cells with the autonomously propagated mCherry-shRNA-HAC were treated with two known aneugens: taxol (paclitaxel) and nocodazole, two HDAC inhibitors: romidepsin and SAHA, the poly(ADP-ribose) polymerase (PARP) inhibitor: olaparib, and the topoisomerase I inhibitor: LMP400.
For each compound, a cell cytotoxicity assay was carried out to determine IC50 values, i.e., the conditions under which the viability of cells would be around 50%. This parameter was chosen in order to normalize the results at the same percentage of viable cells [15]. Time treatments and drug concentrations corresponding to the IC50 are summarized in Table 1. After treatment, the cells were grown for two weeks in the absence of selection. Samples were analyzed every few days, and the proportion of fluorescent cells was determined by FACS. Green-fluorescent cells could be detected after a few days, and treated and untreated cell populations were clearly distinguishable after 5-7 days. The delay between HAC loss and the appearance of non-fluorescent cells is due to the persistence of the shRNA against EGFP. The sampling time had a broad interval (5-14 days) without a significant affect on the calculated rate of HAC loss similar to that previously determined for the original “loss of signal” system [15, 25]. Figure 2B shows representative flow cytometry histograms illustrating a gain of green fluorescence in cell populations treated by a single doze of taxol or gemcitabine. Importantly, the measurements were highly reproducible: the raw FACS data of three independent populations for drug treatments have standard deviations less than 1%.
Table 1. List of compounds used in this study.
| Drugs* | Target | IC50** |
|---|---|---|
| Taxol | Microtubule Stabilizing | 10 nM |
| Nocadazole | Microtubule Destabilizing | 5 μM |
| SAHA | Histone Deacetylase | 2 μM |
| Romidepsin | Histone Deacetylase | 2 nM |
| Olaparib | Poly ADP Ribose Polymerase | 10 μM |
| LMP400 | Topoisomerase I | 50 nM |
| Gemcitabine | Ribonucleotide Reductase | 50 nM |
| PF-2771 | CENP-E | 10 μM |
| GSK923295*** | CENP-E | 10 μM |
| Zoledronic acid | CENP-F | 10 μM |
| siRNA target | Sense sequence as ordered**** |
siRNA per well (6-well plate) |
| SKA3 | 5′-AGACAAACAUGAACAUUAAUU-3′ | 25 pmol |
| CENP-E | 5′-AACACGGAUGCUGGUGACCUC-3′ | 25 pmol |
| MIS18β | 5′-AGGCAGUACUUACAACCUUUU-3′ | 25 pmol |
| ON-TAGET plus, Non-targeting pool | 5′-UGGUUUACAUGUCGACUAA-3′ 5′-UGGUUUACAUGUUGUGUGA-3′ 5′-UGGUUUACAUGUUUUCUGA-3′ 5′-UGGUUUACAUGUUUUCCUA-3′ |
25 pmol |
All drugs, except RF-1771, GSK92395 and zoledronic acid, were described in our previous publications [15, 25]. GSK92395 was purchased from Selleckchem (Catalog no. S7090). PF-1771 was kindly provided by Dr. B.W. Murray (Pfizer Worldwide Research and Development, La Jolla Laboratories, San Diego). Zoledronic acid was purchased from Novartis Pharma AG (Stein, Switzerland).
The cells were treated overnight.
For HT1080 cells containing EGFP-HAC, IC50 for GSK923295 was 2 μM. For HT1080 cells containing mCherry-shRNA-HAC, IC50 was 10 μM.
The two underlined uracil (UU) were added by Dharmacon as part of siRNA construction.
FISH analysis was used to confirm that the appearance of fluorescent cells detected by flow cytometry corresponded to HAC loss (Supplementary Figure S5). Quantitative analysis of metaphase chromosome spreads using a HAC-specific probe (see MATERIALS AND METHODS) correlated with the data on HAC loss determined by FACS (see Supplementary Table S2). Therefore, we conclude that the appearance of green fluorescent cells is caused by HAC loss.
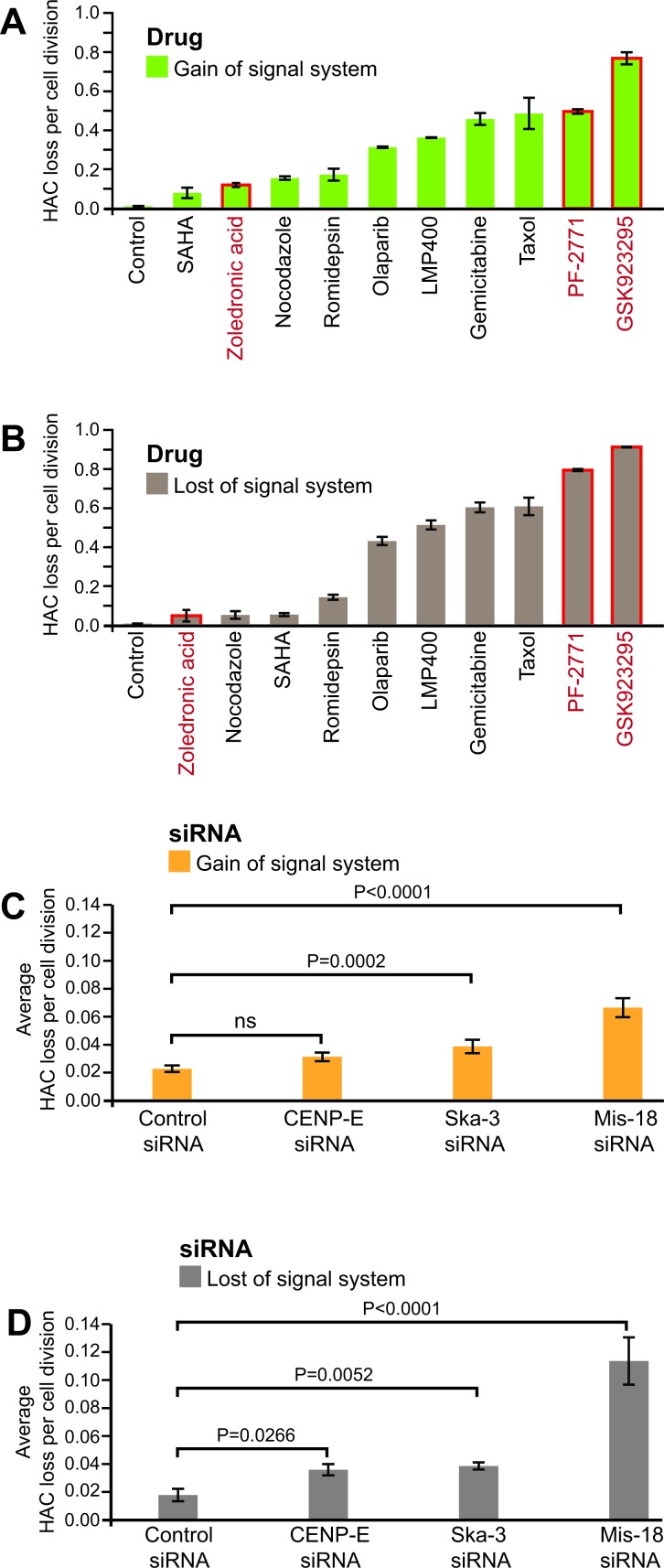
Figure 3A summarizes the estimated rates of HAC loss in response to the analyzed drugs. The rate of HAC loss (RDrug) induced by drug treatment was calculated from the proportion of fluorescent cells in the population (see MATERIALS AND METHODS). The data reveal that the drugs affect the rates of HAC mis-segregation during mitotic divisions differently. The highest effect for this group of established drugs was detected for taxol and gemcitabine. A single dose of these drugs increased the rate of HAC loss ∼40 and 50 fold, respectively. To verify the accuracy of measuring HAC loss using this “gain of signal” system, in parallel experiments the effect of the same compounds was measured using the original “loss of signal” system. The results of this analysis are presented in Figure 3B and clearly demonstrate that ranking the drugs according to their effect on HAC loss using two different systems is indistinguishable.
Figure 3. Measurements of CIN induced through treatment by anticancer agents or gene knockouts.
A., B. Effect of anticancer drugs on CIN was measured by “gain of signal” and “loss of signal” HAC-based assays. HT1080 cells containing either the mCherry-shRNA-HAC or EGFP-HAC were treated by ten different compounds as described in MATERIALS AND METHODS. The rate of HAC loss per cell division was calculated based on the ratio of HAC-positive, HAC-negative cells and the average time per cell division. The drug concentrations used were at IC50 for HT1080 cells. The strongest CIN inducers identified were gemcitabine, taxol, PF-2771 and GSK923295. The control corresponds to the frequency of spontaneous HAC loss in HT1080 cells. C., D. Effect of gene knockouts on the rate of HAC loss. HT1080 cells carrying either the mCherry-shRNA-HAC or EGFP-HAC were treated by different siRNAs and the rate of HAC loss per generation was calculated as described in MATERIALS AND METHODS. In both systems, the strongest CIN effect was observed after cells treatment with MIS18β siRNA. The control corresponds to a frequency of HAC loss after treatment by a negative control siRNA.
Drugs known to increase CIN beyond the therapeutic threshold are currently few in number, and the clinical promise of targeting the CIN phenotype warrants new screening efforts. Taking this in account, in this work we analyzed three new promising anticancer drugs, PF-2771 and GSK923295 [28] that are inhibitors of the centromere-associated protein CENP-E [29]. (CENP-E is a microtubule plus-end-directed kinetochore motor required for congression of pole-proximal chromosomes [30]) and a potential inhibitor of the kinetochore protein CENP-F, zoledronic acid [31]. Therefore, these inhibitors might induce a high rate of chromosome mis-segregation. Indeed, treatment of cells by PF-2771 and GSK923295 increased the rate of HAC loss to values equal to or exceeding those observed after taxol or gemcitabine treatment (Figure 3A and 3B). Note, these two compounds exhibited the highest effect on HAC loss based on a ranking of 62 anticancer compounds [25].
We conclude that this new “gain of signal” assay is suitable for comparing the effect of various drugs on CIN.
Quantitative profiling of CIN induced by gene knockouts
One difficulty in identifying mammalian CIN regulators and determining their relative importance in cancer is the lack of a straightforward quantitative assay for CIN in live mammalian cells. To address this point, we analyzed whether the new “gain of signal” system could be applied for detection of CIN induced by the knockout of genes controlling mitotic progression through distinct mechanisms. We performed the experiments on measuring the HAC stability after siRNA depletion of three proteins: the CENP-A loading cofactor MIS18β, the centromere-associated protein CENP-E and a spindle/kinetochore-associated protein SKA3/RAMA1 [29, 32-35]. As seen in Figure 3C, depletion of MIS18β or SKA3/RAMA1 protein significantly increased the frequency of HAC loss. No detectable destabilization of HAC was observed after cell treatment by control siRNA. Control Western blots showed a decrease of corresponding proteins in the treated cells (Supplementary Figure S6). FISH analysis of metaphase chromosome spreads using a HAC-specific probe correlated with the data on HAC loss determined by FACS (Supplementary Table S2). A similar effect of depletion of the same genes was detected using the original “loss of signal” system (Figure 3D). Collectively, the above results validate the ability of both HAC-based assays to be employed for identification of novel genes controlling chromosome transmission in human cells.
Quantitative profiling of CIN inducing drugs by a fluorescence microplate reader
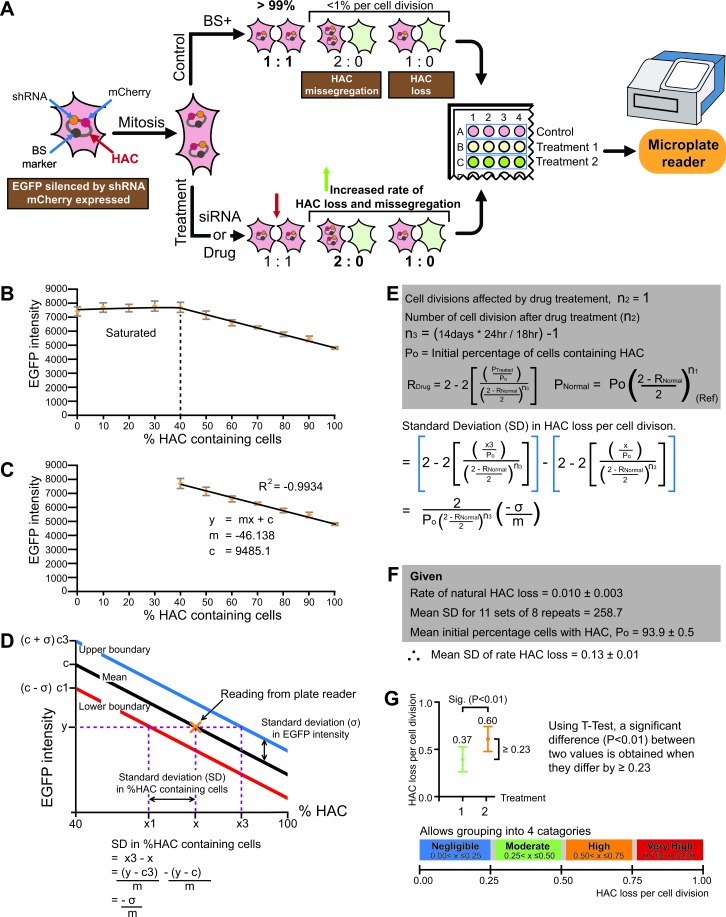
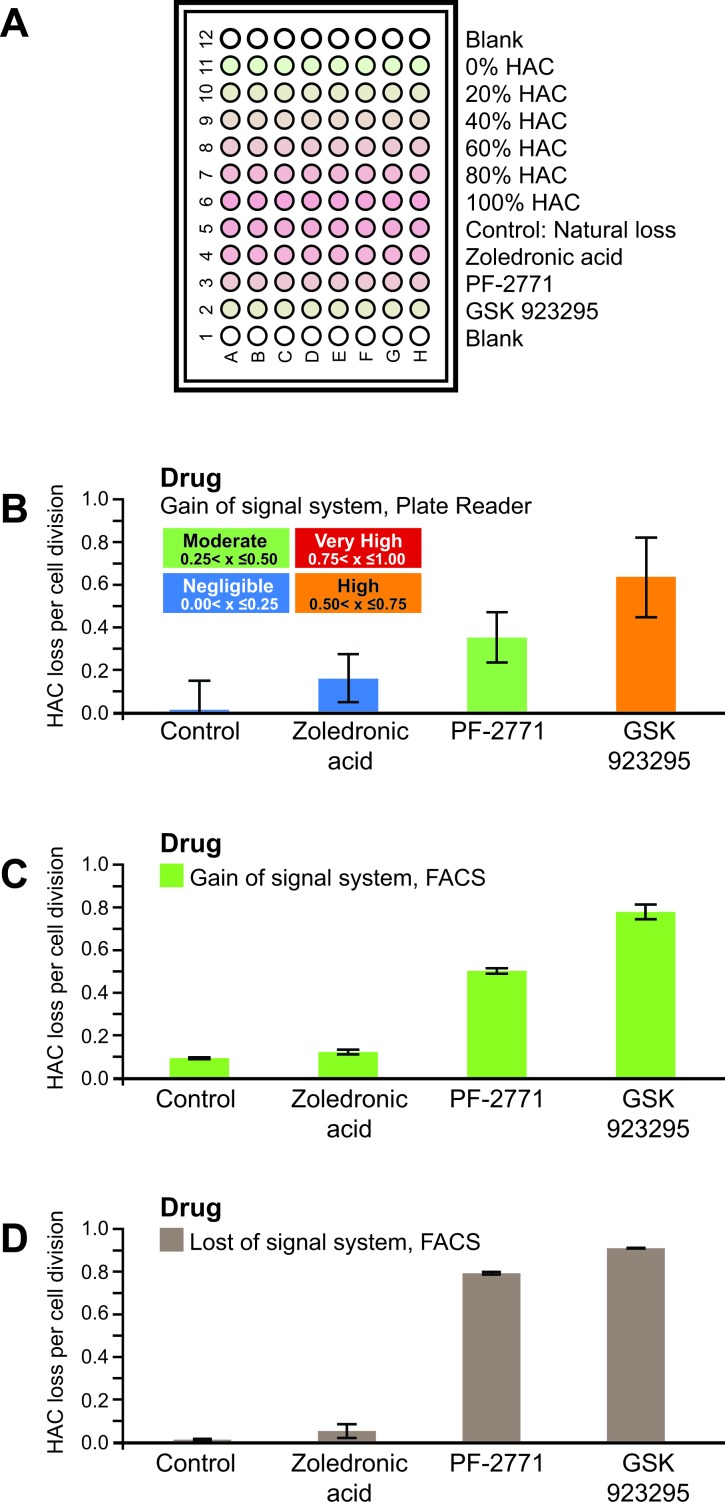
In order to efficiently search chemical or siRNA libraries for drugs or gene knockouts inducing CIN, it is necessary to develop a high-throughput screen (Figure 4A). To that end, we performed a set of pilot experiments. First, we demonstrated that it is possible to construct a titter curve using microplate reader data of EGFP fluorescence from populations of known composition of HAC-containing (not green) and HAC-less (green) cells. A titter curve was generated by mixing two cell populations, the HT1080-shGFP-4 cell line which contains the HAC (not green), and the HT1080-JH1 expressing EGFP and without HAC (green) in varying compositions ranging from 0% to 100%. The green fluorescence of these mixed populations was determined with a microplate reader (Figure 4B and 4C). Using the standard deviation of the plate reader data, we found that a significant difference at (P < 0.01) could be detected if the rate of HAC loss per cell division of two drug treatments differed by 0.23 (Figure 4D-4F) (see for details MATERIAL AND METHODS). This resolution is insufficient to rank drugs against one another. However, it is sufficient to grade candidate CIN drugs into 4 broad categories (Figure 4G). The standard deviation in the calculated rate of HAC loss was found to be proportional to . Thus, better resolution between candidate drugs can be obtained by either decreasing the standard deviation (σ) of the microplate readings or by increasing the difference in green fluorescence intensity between HAC-less cells and HAC-containing cells (m).
Figure 4.
A. Scheme of a high throughput assay for measuring chromosome instability (CIN). During S-phase the HAC is duplicated in the cell as other host chromosomes. Under normal growth conditions (control), after cell division the majority of cells in population contain the HAC (1:1 segregation). After drug or siRNA treatment, one copy of HAC can be lost during cell division (1:0 segregation) or two copies of HAC can mis-segregate (2:0 segregation). The cells that inherited the HAC are fluorescent red while cells that lost it are fluorescent green. For each treatment, the average fluorescence of each cell sample may be measured using a multi-well plate reader. Thus, the genes involved in chromosome transmission or drugs affecting chromosome stability may be identified. B. An example of a titration curve between EGFP intensity against cell populations of varying percentage of HAC-containing (not green) and HAC-less (green) cells obtained from a fluorescence microtiter plate reader. C. Construction of a linear curve between a cell population's EGFP intensity against the percentage of HAC-containing cells. D. The relationship between the standard deviation of a cell population's EGFP intensity and it's percentage of HAC-containing cells. E. The relationship between the standard deviation of HAC-containing cells and the ‘rate of HAC loss per cell division'. F. The estimated standard deviation in the ‘rate of HAC loss' using data derived from a fluorescence plate reader and known rates of natural HAC loss. G. The minimum difference between two values of ‘HAC loss' needed to be significant (P < 0.01) was calculated using a T-test and the standard deviations derived above. The results indicate that plate reader is only able to discriminate drugs into broad categories.
Then we analyzed three anticancer drugs, PF-2771, GSK923295 and zoledronic acid, using a microplate reader. As seen from Figure 5, treatment of HAC-containing cells by PF-2771 or GSK923295 experienced a dramatic increase in the rates of HAC loss (Figure 5C and 5D) that could be quantified by a fluorescence microplate reader (Figure 5B). Thus, the “gain of signal assay” may be adapted to a 96-well microplate format and used to detect the fluorescence after treatment by a compound.
Figure 5.
A. An illustration of how a 96-well plate was loaded with drug treated cells. Populations of known mixtures of HT1080-shGFP-4 (HAC+) and HT1080-JH1 (HAC-) cells were included as a ladder. B. The calculated rates of HAC loss per cell division for three drugs, zoledronic acid, PF-2771 and GSK923295, using the “gain of signal” system with a fluorescence microtitter plate reader. C. The calculated rates of HAC loss for the same drugs using the “gain of signal” system with FACS. D. The calculated rates of HAC loss for the same drugs using the ”loss of signal' system with FACS.
DISCUSSION
Most solid tumors are aneuploid, carrying an abnormal number of chromosomes, and they frequently mis-segregate whole chromosomes in a phenomenon termed chromosome instability (CIN). Despite its importance in cancer etiology and therapy, until recently there were no fast, straightforward, quantitative assays for CIN. To address this problem, previously we developed quantitative CIN assay based on a Human Artificial Chromosome (HAC) carrying a constitutively expressed EGFP transgene [15]. In this assay, HAC loss events induced by drug treatment could be measured as the fraction of cells that lose EGFP fluorescence signal using routine flow cytometry. This “loss of signal” assay is simple and reproducible and it was successfully used to rank a set of different compounds for their effect on CIN [25]. Unfortunately, this assay is not suitable for the development of a high-throughput screening method. It is difficult to detect the loss of EGFP fluorescence in a few cells against a bright background of EGFP-positive cells. However the opposite, the gain of EGFP fluorescence against a background of colorless cells is much easier to detect. Thus, an assay based on “gain of signal” would potentially be more suitable for high-throughput screening and perhaps earlier detection of the induced CIN phenotype.
In this report, we have described the development and validation of a novel quantitative HAC-based assay for assessing chromosome stability in human cells. A key design feature of this assay is that the fluorescence signal is gained in the cells after loss of a HAC that carries a constitutively expressed shRNA against an EGFP transgene integrated elsewhere into the human genome. Thus, cells that inherit the HAC do not display green fluorescence, while cells lacking the HAC do. To verify the accuracy of this new assay, in parallel experiments we measured the rates of HAC loss in a response to cells treatment by a set of anticancer drugs using “gain of signal” and “loss of signal” systems with flow cytometry as a read-out. The relative ranking of drugs that strongly induced CIN did not change, although the same could not be said for drugs that weakly induced CIN. As the aims of this system was to identify drugs that strongly induced CIN, the new “gain of signal” assay may be used both for ranking drugs on their effect on CIN and also for identifying new compounds that promote CIN. During these experiments we identified two drugs, i.e. the newly developed inhibitors of the centromere-associated protein CENP-E, PF-2771 and GSK923295, which induce an extremely high rate of HAC loss. These compounds along with several other drugs that greatly increase CIN, including paclitaxel, olaparib, talazoparib, LMP400 and gemcitabine [25], may be considered as a first choice when CIN is considered as a target for cancer therapy.
In addition, we demonstrated that both HAC-based systems, i.e. “loss of signal” and “gain of signal”, are sensitive enough for detection of CIN induced by depletion of genes, for example, controlling mitotic progression through distinct mechanisms. We demonstrated that siRNA depletion of two genes, MIS18β and SKA3/RAMA1, was accompanied by HAC destabilization to a greater or lesser extent. At present, approximately 400 human genes which control transmission of chromosomes have been annotated with gene ontology (GO) terms, while systematic CIN gene screens in yeast have revealed 692 genes [5]. Therefore, in the future these HAC-based assays may be useful in genome-wide screening to uncover novel CIN genes in humans. The identification of a comprehensive set of genes controlling chromosome segregation should expedite the development of new therapeutic strategies to target the CIN phenotype of cancer cells.
We also examined the potential of the HAC-based “gain of signal” assay in a high-throughput screen of chemical libraries and identification of new genes that modulate CIN (e.g. using multiple gene depletion screens). The pilot experiments were performed where we combined the assay with a 96-well fluorescence microplate reader (see Figures 4 and 5). Although the fluorescence microplate reader was far less sensitive than the flow cytometer, we were able to broadly quantify CIN inducing drugs into 4 categories (Negligible, Moderate, High and Very High) and quantitatively distinguish the drugs by a fluorescence microplate reader.
It is worth noting that this method has some limitations due to the fluorescence signals detection. We expect to improve the present assay by: i) increasing the expression of the EGFP transgene by generating new HAC-host cell lines with EGFP integrated into different chromosomal sites; ii) expressing multiple shRNA targeted to different parts of the EGFP mRNA and, thus, improving silencing; iii) reengineering the mCherry to be constitutively expressed and act as a marker for cell density. In addition, the sensitivity of the assay may be significantly improved using another plate reader system for detection of a fluorescence signal. This work is currently in progress. Also, the current labeling of the HAC does not allow us to simply distinguish between HAC loss (1:0) and HAC mis-segregation (2:0) events. (It is possible only by using a time-consuming interphase FISH). In principle, this limitation may be overcome by expression of the tetracycline repressor fused to mCherry (TetR-mCherry). In such cells the alphoidtetO-HAC may be monitored the live following TetR-mCherry signal. Alternative may be loading a copy number dependent, selectable marker (e.g. the dihydrofolate reductase gene) into the HAC where an increased copy number of this gene is accompanied by a higher resistance of cells to methotrexate.
To summarize, a novel “gain of signal” HAC-based assay allows straightforward, quantitative assessment of CIN under a variety of conditions. The assay will be employed to identify novel agents that specifically elevate chromosome mis-segregation and drive lethal aneuploidy. The assay is also suitable for synthetic lethal screens in which combinatorial effects of disruption of two or more pathways can be studied. In future, this “gain of signal” assay may be adapted for high-throughput chemical screens using a fluorescence microplate reader to characterize chemical libraries and identify new conditions that modulate CIN levels, thereby laying the foundation for new treatment strategies for cancer.
MATERIALS AND METHODS
Cell lines and culture
Human fibrosarcoma HT1080 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% (v/v) tet system-approved fetal bovine serum (Clontech Laboratories, Inc.) at 37°C in 5% CO2. Hypoxanthine phosphoribosyltransferase (HPRT)-deficient Chinese hamster ovary (CHO) cells (JCRB0218) carrying the alphoidtetO-HAC [22, 24, 25] were maintained in Ham's F-12 nutrient mixture (Invitrogen) plus 10% FBS with 8 μg/ml of BS (Funakoshi). After loading of the mCherry-shRNA cassette (see below) into the alphoidtetO-HAC, the CHO cells were cultured in 1× HAT supplemented medium.
Construction of the tDNA- mCherry- shRNA plasmid (A245)
Scheme of construction of the A245 plasmid is presented in Supplementary Figure S2. This plasmid was constructed in three steps. In step 1, the HuSH pGFP-V-RS plasmid (OriGene) was digested with BamHI /HindIII and the shGFP sequence was ligated in as a double stranded oligo formed by annealing primers J001 and J002. The plasmid created was HuSH-pRS-shGFP. In step 2, the pcDNA 3.1(+) (Thermofisher Scientific) was digested with HindIII and XbaI. Then the mCherry fragment was PCR amplified from the pRSET-B-mCherry plasmid (Tsien Lab, University of California, San Diego) using the primers J005 and J006, digested with HindIII and XbaI. The two fragments were ligated, creating the plasmid pcDNA-mCherry. In step 3, the A245 plasmid was completed in a 3-way ligation of the following fragments: the U6-shGFP fragment, obtained by PCR amplification of HuSH-pRS-shGFP plasmid using the primers J003 and J004, followed by digestion with SpeI and XhoI; the tDNA-loxP-3′HPRT fragment, obtained by SpeI / BamHI digest of the CAG-EGFP-tDNA plasmid [26]; and lastly the CMV-mCherry-BGHpA fragment which was obtained by PCR amplification of pcDNA-mCherry using the primers J007 and J008, followed by digestion with XhoI and BamHI. Sequences of all primers used are in Supplementary Table S1.
Construction of the tDNA-EGFP-HyTK plasmid (A158)
Scheme of construction of the plasmid A158 is presented in Supplementary Figure S4. The plasmid A158 was constructed in three steps. In step 1, the plasmid A154 was constructed in a 3-way ligation of three fragments: the tDNA-CAG fragment, obtained from EcoRI/SacI digestion of the CAG-EGFP-tDNA plasmid [26]; the pBS backbone, obtained from SacI / BamHI digestion of pBlueScript II KS (Stratagene); and the EGFP fragment, obtained by PCR amplification of the plasmid p264 [22] using the primers B119/B052, followed by BamHI/EcoRI digestion. In step 2, the plasmid A156 was constructed in a 3-way ligation of three fragments: the T2A-HyTK fragment, obtained from BamHI /XhoI digestion of the CAG-attB-HyTK plasmid [36]; the tDNA-P fragment, obtained by PCR amplification of pCR4-TOPO-tDNA [27] using the primers B405 / B404 followed by SalI / NotI digestion; and the pBS backbone, obtained by NotI /BamHI digestion of pBlueScript II KS (Stratagene). In step3, the A158 plasmid was completed in a 3-way ligation of the following three fragments: the tDNA-CAG-EGFP fragment was obtained by SacI /BamHI digestion of the plasmid A154; the T2A-HyTK-tDNA fragment was obtained by BamHI / NotI digestion of the A156 plasmid; and the pBS backbone was obtained by PCR amplification of pBlueScript II KS (Stratagene) using the primers B396/B395 followed by digestion with NotI / SacI. Sequences of all primers used are in Supplementary Table S1.
Loading of the tDNA-mCherry-shRNA plasmid into the loxP site of alphoidtetO-HAC in hamster CHO cells
3 μg of the A245 plasmid and 1 μg of the Cre expression pCpG-iCre vector DNA were co-transformed into HPRT-deficient hamster CHO cells containing the alphoidtetO-HAC by lipofection with FuGENEHD transfection reagent (Roche) or Lipofectamine 2000 (Invitrogen). HPRT-positive colonies were selected after 2 to 3 weeks growth in HAT medium. For each experiment, from 5 to 7 clones were usually selected. Correct loading of the A245 plasmid into the HAC was confirmed by genomic PCR [26] with a specific pair of primers that diagnose reconstitution of the HPRT gene (Supplementary Table S1).
Generation of the EGFP-expressing human cell line
The EGFP-containing plasmid A158 was linearized with ScaI and transfected into HT1080 cells. Transfectants were selected on DMEM plus FBS medium with 80 μg /ml Hygromycin B (Invitrogen, USA). Of the colonies obtained, the clone HT1080-JH1 exhibited the strongest expression of EGFP was chosen for further experiments to host the HAC that constitutively expressed shRNA against EGFP.
Microcell-mediated chromosome transfer
The alphoidtetO-HAC containing the mCherry-shRNA cassette was transferred from hamster CHO cells to the human HT1080-JH1 cell line using a standard microcell-mediated chromosome transfer (MMCT) protocol [22, 37]. 12 μg/ml blasticidin (BS) was used to select resistant colonies containing the HAC. Typically, three to ten BSR colonies are obtained in one MMCT experiment. BSR colonies were analyzed by FISH for the presence of the autonomous form of the HAC. The potential co-transfer of CHO chromosomes was excluded using a sensitive PCR test for rodent-specific SINE elements (Supplementary Table S1). The clone HT1080-shGFP-4 was selected for further work and was maintained under selection with 12 μg/ml blasticidin and 80 μg/ml hygromycin B.
Flow cytometry
Analysis of EGFP expression was performed using LSRFortessa (BD Biosciences) flow cyctometer and BD FACSDiva software [15]. The cells were harvested by trypsin-treatment. Intensities of fluorescence were determined by flow cytometry. 1.0 × 104 cells were analyzed for each cell sample.
Drug treatment
Ten different drugs were used in our experiments (Table 1). The experiment protocol was as follows. HT1080 cells containing EGFP-HAC were maintained on 6 μg/ml blasticidin selection while HT1080 cells containing mCherry-shRNA-HAC was maintained on 12 μg/ml blasticidin and 80 μg/ml hygromycin B. Approximately 1 × 105 cells were cultured either in the presence or absence of blasticidin selection in parallel with a third culture that was exposed to the agent under examination to test its effect on HAC segregation. The drug concentration applied was adjusted to the IC50 level for each compound which was determined using a proliferation assay described below. Concentrations of drugs and lengths of treatment are presented in Table 1. Subsequently, the drug was removed by performing three consecutive medium washes and the cells were subsequently grown without antibiotic selection for 13 days. At the end of the experiment, cells were collected and analyzed by flow cytometry to detect the proportion of cells that gain or loss EGFP fluorescence. This served as a measure of HAC stability following drug treatment. For all drugs, experiments were carried out in triplicate.
Treatment by siRNAs
SKA3/RAMA1, MIS18β and CENP-E were depleted using established siRNAs targeting published sequences [29, 32-34]. siRNAs used to target SKA3, CENP-E and MIS18β are listed in Table 1. siRNAs were purchased from Dharmacon (Lafayette, CO). For siRNA treatment, 1×105 / well cells were seeded in 6 well plates before a day of the experiment. Cells were transfected with each siRNAs using lipofectamine-RNAiMAX (Invitrogen). Cells were grown without blasticidine S/hygromycin B selection for 14 days. Silencing efficiency of each protein was monitored by Western blot analysis. On Day 14, cells were collected and analyzed by flow cytometry to detect the proportion of cells that reactivated EGFP fluorescence or lost EGFP signal. All experiments were carried out in triplicate.
Calculation of the drug induced rate of HAC loss
To calculate the rate of HAC loss per cell division after cell treatment by a single dose of drug, we used the equations;
| Eq1 |
[15]
| Eq2 |
[15]
RNormal is the natural rate of HAC loss per cell division. RDrug is the drug induced rate of HAC loss per cell division. P0 is the percentage of EGFP(+) cells when cultured under HAC selection. PNormal is the percentage EGFP(+) cells when cells are cultured without selection. n1 = the number of cell doublings that occurred during culturing without selection. PTreated is the percentage of EGFP(+) cells at the end of the experiments. n1 is the number of cell doublings that occurs during drug treatment. As drug treatment was limited to less than 18 hrs, drug induced HAC loss was limited to a single cell cycle, n1 = 1. n2 is the number of cell doublings that occurs during the culturing without selection after the drug treatment. The doubling time of HT1080 is approximately 18 hrs.
Calculation of the siRNA induced rate of HAC loss
We were unable to quantify the number of cell divisions that siRNA knockdown impacted. Hence, we chose to calculate the average rate of HAC loss per cell division for entire duration of the siRNA experiment. The equation used was . PN is the percentage of EGFP(+) cells at the end of the experiments. n4 is the number of cell doublings that occurs during the experiment. We estimated this value by counting the size of the cell population at three time points. The first time point was on day the wells were seeded. The second time point was on Day 3, 48 hrs after siRNA transformation. After which, 10% of these cells were plated onto a 75cm3 cell culture flask and left to grow until Day 14. The third time point was on Day 14. Cell counts were made using a hemocytometer and viability was determined by 5 min staining with DRAQ7 (Adcam) at concentration stated by the manufacturer and flow cytometry.
Cell viability test
MTS tetrazolium cell viability assays were done according to the manufacturer's instructions (CellTiter 96 AQueous Assay reagent; Promega). Briefly, the CellTiter 96 AQueous One Solution Reagent was added to each well and incubated at 37°C for 3 hr. Cell proliferation was determined by measuring the absorbance at 490 nm using a microplate reader (Molecular Devices). The half-maximal inhibitory concentration (IC50) was obtained from the MTS viability curves using GraphPad Prism 5. Experiments were carried out in triplicate.
FISH analysis with the BAC probe
HT1080 cells were processed for fluorescence in situ hybridization (FISH) after drug treatment followed by the 14 day washout. The probe used for FISH was BAC32-2-mer(tetO) DNA containing 40 kb of alphoid-tetO array cloned into a BAC vector as described previously [16]. Specifically, a BAC32-2-mer(tetO) clone contains an amplified synthetic alphoid DNA dimer. One monomer of this dimer is an alphoid DNA consensus sequence carrying the tetO sequence; another monomer is alphoid DNA from chromosome 17. This probe is specific to the HAC but also gives a low signal with centromeric regions of several endogenous chromosomes. BAC DNA was digoxigenin-labeled using a nick-translation kit with digoxigenin-11dUTP or biotin16-dUTP (Roche Diagnostics). Images were captured as before. The probe was denatured for 5 min at 95°C and added to the slides, which were incubated at 72°C for 2 min before overnight incubation at 39°C. After washes with 0.1 × SSC at 65°C followed by a wash with 4 × SSC + 0.1% Tween 20 at room temperature, standard procedures were used to detect biotinylated probes. Slides were mounted with VectaShield and screened for the presence or absence of the HAC. Between 70-150 metaphases were analyzed for each drug treatment.
Genomic DNA preparation and PCR analysis
Genomic DNA for PCR analysis was prepared using a QIAmp DNA Mini Kit (QIAGEN). Reconstitution of the HPRT gene after Cre/lox-mediated recombination was determined by specific primers Lox137-R, Rev#6 and SV40 PA term rev (Supplementary Table S1). Cross contamination by hamster chromosomes after HAC MMCT transfer from hamster cells to human cells was determined by specific primers detecting hamster SINEs: Cons B2-F, Ham B2-F and Ham B2-R (Supplementary Table S1). PCR products for sequencing were separated by agarose gel electrophoresis and gel extracted.
Western blot analysis
siRNA depletion of proteins was confirmed by Western blot. The following antibodies were used for immunoblotting: Anti-OIP5 (MIS18β) antibody (ab168516), Anti-SKA3 antibody (ab175951), Anti-GAPDH antibody (Cell Signaling).
Fluorescence microplate reader
20,000 cells / well were plated onto a 96 well, treated, flat bottom, tissue culture imaging plate, (BD falcon, REF 353219) and left to grow overnight with 80 μg/ml hygromycin B. On the next day, complete confluence was achieved, and the cells cultures were washed twice with PBS (Life Technologies) with 0.9 mM CaCl2 and 0.5 mM MgCl2. Cells were then lysed with 50 μl of CelLytic Sigma (C2978-50ml) and immediately read with a fluorescence microplate reader (Synerg HT Biotech) (Figure 4A)
Calculation of the rate of HAC loss using a microplate reader
Two cell populations, the HT1080-shGFP-4 cell line which contains the HAC (not green) and the HT1080-JH1 expressing EGFP and without HAC (green) were mixed in varying compositions ranging from 0% to 100%. A titter curve was generated. The green fluorescence of these mixed populations was determined with a microplate reader (Figure 4B). The experiment was repeated three times, each with 8 replicates. The titration generated a linear curve between EGFP intensity and cell populations that ranged from 40% to 100% HAC (R2 = −0.9934) (Figure 4C). Below 40% HAC-containing cells in population, the plate reader was saturated at the settings used.
The average standard deviation from microplate reader readings was determined and used to generate two parallel lines, one standard deviation above and below the mean linear curve. As illustrated in Figure 4D, these two curves were used to determine the standard deviation in the percentage of green fluorescence cells. Then equations Eq1 and Eq2 were used to derive the standard deviation of the rate of HAC loss per cell division (Figure 4F). T-test was then used to determined how large a difference between two values is required to obtain a significant difference at P < 0.01 (GraphPad Prism 5) (Figure 4G).
In addition, by plugging in the known rate of natural HAC loss in HT1080, the starting percentage of HAC containing cells and all possible value of %HAC in a population into Eq1 and Eq2, we determined that the lowest percentage of HAC-containing cells for a single drug dose was 42%. This is within the detection limit of plate reader at the settings used.
SUPPLEMENTARY MATERIAL FIGURES AND TABLES
Acknowledgments
PF-2771 was kindly provided by Dr. B.W. Murray (Pfizer Worldwide Research and Development, La Jolla Laboratories, San Diego). The authors also would like to thank the CRC, LRBGE Fluorescence Imaging Facility (NIH) and personally Dr. Karpova and Dr. McNally for instructions, consultations and help with the usage of a DeltaVision microscopy imaging system. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, USA (VL), a Wellcome Trust Principal Research Fellowship [grant number 073915] (WCE), MEXT KAKENHI [grant number 23114008] (HM) and the Kazusa DNA Research Institute Foundation (HM).
Footnotes
Authors' contribution
Vladimir Larionov and Jung-Hyun Kim conceptualized and designed experiments. Vladimir Larionov, Natalay Kouprina and Nicholas Lee wrote the manuscript. Jung-Hyun Kim, Hee-Sheung Lee, Nicholas Lee and Nikolay Goncharov performed the experiments. Vadim Kumeiko supported the study. Hiroshi Masumoto and William Earnshaw supported the study and revised critically the article for important intellectual content.
CONFLICTS OF INTERESTS
The authors have no conflicts of interests to disclose.
REFERENCES
- 1.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Current Biology. 2010;20:R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 2015:10. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, McGranahan N, Dewhurst SM, Burrell RA, Tomlinson I, Swanton C. Cancer: Evolution Within a Lifetime. Annu Rev Genet. 2014;48:215–236. doi: 10.1146/annurev-genet-120213-092314. [DOI] [PubMed] [Google Scholar]
- 4.Lee H. How Chromosome Mis-Segregation Leads to Cancer: Lessons from BubR1 Mouse Models. Mol Cells. 2014;37:713–718. doi: 10.14348/molcells.2014.0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stirling PC, Bloom MS, Solanki-Patil T, Smith S, Sipahimalani P, Li Z, Kofoed M, Ben-Aroya S, Myung K, Hieter P. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genetics. 2011;7:e1002057. doi: 10.1371/journal.pgen.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssen A, Kops GJPL, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li QY, Hardcastle T, Lee A, Roy R, East P, Kschischo M, Endesfelder D, Wylie P, Kim SN, Chen JG, Howell M, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci USA. 2009;106:8671–8676. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colombo R, Moll J. Targeting aneuploid cancer cells. Expert Opinion on Therapeutic Targets. 2011;15:595–608. doi: 10.1517/14728222.2011.558007. [DOI] [PubMed] [Google Scholar]
- 9.Janssen A, Kops GJ, Medema RH. Targeting the mitotic checkpoint to kill tumor cells. Hormones & Cancer. 2011;2:113–116. doi: 10.1007/s12672-010-0059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen A, van der Burg M, Szuhai K, Kops GJPL, Medema RH. Chromosome Segregation Errors as a Cause of DNA Damage and Structural Chromosome Aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 11.Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci U S A. 2013;110:E4134–4141. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirsch-Volders M, Elhajouji A, Cundari E, Van Hummelen P. The in vitro micronucleus test: a multi-endpoint assay to detect simultaneously mitotic delay, apoptosis, chromosome breakage, chromosome loss and non-disjunction. Mutation Research. 1997;392:19–30. doi: 10.1016/s0165-1218(97)00042-6. [DOI] [PubMed] [Google Scholar]
- 13.Bakker B, van den Bos H, Lansdorp PM, Foijer F. How to count chromosomes in a cell: An overview of current and novel technologies. Bioessays. 2015;37:570–577. doi: 10.1002/bies.201400218. [DOI] [PubMed] [Google Scholar]
- 14.Thompson LL, McManus KJ. A Novel Multiplexed, Image-Based Approach to Detect Phenotypes That Underlie Chromosome Instability in Human Cells. PLoS One. 2015:10. doi: 10.1371/journal.pone.0123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HS, Lee NCO, Grimes BR, Samoshkin A, Kononenko AV, Bansal R, Masumoto H, Earnshaw WC, Kouprina N, Larionov V. A new assay for measuring chromosome instability (CIN) and identification of drugs that elevate CIN in cancer cells. BMC Cancer. 2013;13:252. doi: 10.1186/1471-2407-13-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Developmental Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouprina N, Earnshaw WC, Masumoto H, Larionov V. A new generation of human artificial chromosomes for functional genomics and gene therapy. Cellular and Molecular Life Sciences. 2013;70:1135–1148. doi: 10.1007/s00018-012-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshimura M, Uno N, Kazuki Y, Katoh M, Inoue T. A pathway from chromosome transfer to engineering resulting in human and mouse artificial chromosomes for a variety of applications to bio-medical challenges. Chromosome Res. 2015;23:111–133. doi: 10.1007/s10577-014-9459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouprina N, Tomilin AN, Masumoto H, Earnshaw WC, Larionov V. Human artificial chromosome-based gene delivery vectors for biomedicine and biotechnology. Expert Opinion on Drug Delivery. 2014;11:517–535. doi: 10.1517/17425247.2014.882314. [DOI] [PubMed] [Google Scholar]
- 20.Katona RL. De novo formed satellite DNA-based mammalian artificial chromosomes and their possible applications. Chromosome Res. 2015;23:143–157. doi: 10.1007/s10577-014-9458-0. [DOI] [PubMed] [Google Scholar]
- 21.Kim JH, Kononenko A, Erliandri I, Kim TA, Nakano M, Iida Y, Barrett JC, Oshimura M, Masumoto H, Earnshaw WC, Larionov V, Kouprina N. Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc Natl Acad Sci USA. 2011;108:20048–20053. doi: 10.1073/pnas.1114483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iida Y, Kim JH, Kazuki Y, Hoshiya H, Takiguchi M, Hayashi M, Erliandri I, Lee HS, Samoshkin A, Masumoto H, Earnshaw WC, Kouprina N, Larionov V, Oshimura M. Human artificial chromosome with a conditional centromere for gene delivery and gene expression. DNA Research. 2010;17:293–301. doi: 10.1093/dnares/dsq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kononenko AV, Bansal R, Lee NCO, Grimes BR, Masumoto H, Earnshaw WC, Larionov V, Kouprina N. A portable BRCA1-HAC (human artificial chromosome) module for analysis of BRCA1 tumor suppressor function. Nucleic Acids Res. 2014:42. doi: 10.1093/nar/gku870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouprina N, Samoshkin A, Erliandri I, Nakano M, Lee HS, Fu HG, Iida Y, Aladjem M, Oshimura M, Masumoto H, Earnshaw WC, Larionov V. Organization of Synthetic Alphoid DNA Array in Human Artificial Chromosome (HAC) with a Conditional Centromere. Acs Synth Biol. 2012;1:590–601. doi: 10.1021/sb3000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HS, Lee NC, Kouprina N, Kim JH, Kagansky A, Bates S, Trepel JB, Pommier Y, Sackett D, Larionov V. Effects of Anticancer Drug on Chromosome Instability (CIN) and New Clinical Implications for Tumor-Suppressing Therapies. Cancer Research. 2016;76:902–11. doi: 10.1158/0008-5472.CAN-15-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee NC, Kononenko AV, Lee HS, Tolkunova EN, Liskovykh MA, Masumoto H, Earnshaw WC, Tomilin AN, Larionov V, Kouprina N. Protecting a transgene expression from the HAC-based vector by different chromatin insulators. Cellular and Molecular Life Sciences. 2013;70:3723–3737. doi: 10.1007/s00018-013-1362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebersole T, Kim JH, Samoshkin A, Kouprina N, Pavlicek A, White RJ, Larionov V. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle. 2011;10:2779–2791. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett A, Bechi B, Tighe A, Thompson S, Procter DJ, Taylor SS. Cenp-E inhibitor GSK923295: Novel synthetic route and use as a tool to generate aneuploidy. Oncotarget. 2015;6:20921–20932. doi: 10.18632/oncotarget.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kung PP, Martinez R, Zhu Z, Zager M, Blasina A, Rymer I, Hallin J, Xu M, Carroll C, Chionis J, Wells P, Kozminski K, Fan J, Guicherit O, Huang B, Cui M, et al. Chemogenetic evaluation of the mitotic kinesin CENP-E reveals a critical role in triple-negative breast cancer. Molecular Cancer Therapeutics. 2014;13:2104–2115. doi: 10.1158/1535-7163.MCT-14-0083-T. [DOI] [PubMed] [Google Scholar]
- 30.Barisic M, Sousa RSE, Tripathy SK, Magiera MM, Zaytsev AV, Pereira AL, Janke C, Grishchuk EL, Maiato H. Microtubule detyrosination guides chromosomes during mitosis. Science. 2015;348:799–803. doi: 10.1126/science.aaa5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown HK, Ottewell PD, Coleman RE, Holen I. The kinetochore protein Cenp-F is a potential novel target for zoledronic acid in breast cancer cells. J Cell Mol Med. 2011;15:501–513. doi: 10.1111/j.1582-4934.2009.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci. 2009;122:2436–2445. doi: 10.1242/jcs.051912. [DOI] [PubMed] [Google Scholar]
- 33.Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, Fang J, Zang J, Cheng J, Teng M, Ding X, Yao X. Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP) The Journal of Biological Chemistry. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanudji M, Shoemaker J, L'Italien L, Russell L, Chin G, Schebye XM. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol Biol Cell. 2004;15:3771–3781. doi: 10.1091/mbc.E03-07-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z, Lee NC, Dafhnis-Calas F, Malla S, Smith MC, Brown WR. Site-specific recombination in Schizosaccharomyces pombe and systematic assembly of a 400kb transgene array in mammalian cells using the integrase of Streptomyces phage phiBT1. Nucleic Acids Res. 2008;36:e9. doi: 10.1093/nar/gkm1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koi M, Shimizu M, Morita H, Yamada H, Oshimura M. Construction of mouse A9 clones containing a single human chromosome tagged with neomycin-resistance gene via microcell fusion. Japanese Journal of Cancer Research. 1989;80:413–418. doi: 10.1111/j.1349-7006.1989.tb02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.