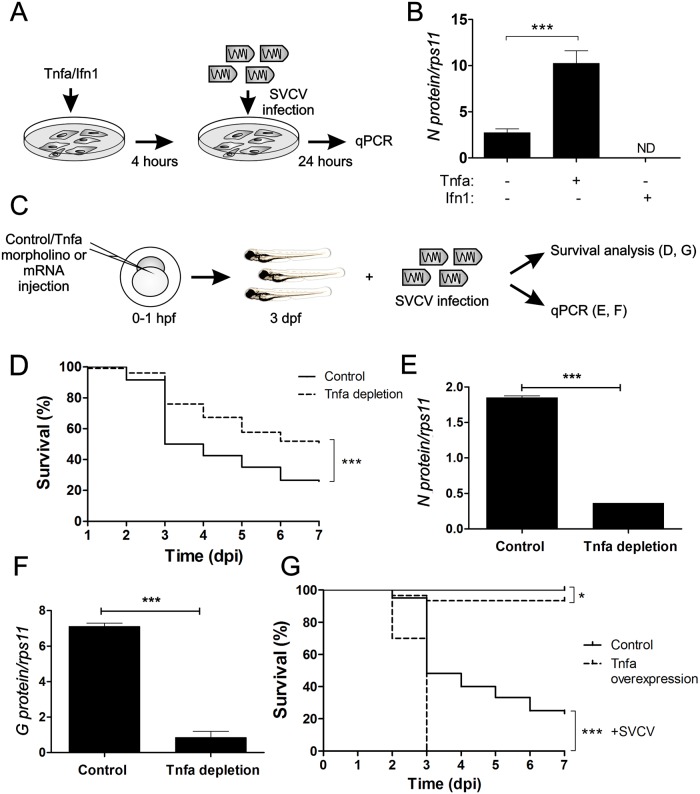
Fig 1. Tnfa enhances SVCV infection in zebrafish.
(A) Workflow of the experimental design followed in (B). Recombinant zebrafish Tnfa or interferon 1 (Ifn1) were added to ZF4 cells growing in monolayer at 80% confluence and incubated for 4 hours. Subsequently, the medium was washed out and fresh medium containing SVCV was added. After 24 hours of incubation with the virus, the cells were harvested for qPCR analysis. (B) N protein mRNA expression levels assessed by qPCR relative to the housekeeping gene rps11 and multiplied by 105. Bars represent mean ± S.E.M. of indicated gene expression from one representative experiment. (C) Workflow of the experimental design followed in (D-G). Std (Control) or Tnfa mos (D-F) or antisense or Tnfa RNAs (G) were injected in zebrafish embryos at one-cell-stage of development. At 3 dpf, these larvae were immerse in RPMI containing inactivated SVCV (control) or intact SVCV for subsequently analysis of survival (D, G) or qPCR analysis at 48 hours post-infection (hpi) (E, F). Percentage of survival of Tnfa-depleted (D) and overexpressing (G) zebrafish larvae exposed to 109 TCID50/ml SVCV. (E, F) The mRNA levels of the gene coding for the SVCV N protein as an estimation of the viral replication (E), and the RNA- levels of G protein (F) were determined in the infected larvae by qPCR in 10 pooled larvae at 48 hpi (5 dpf). The gene expression was normalized against rps11 and multiplied by 105 for N protein. Bars represent mean ± S.E.M. of triplicate readings from pooled larvae and the data are representative of two independent experiments. ***p<0.001. ND, not detected.