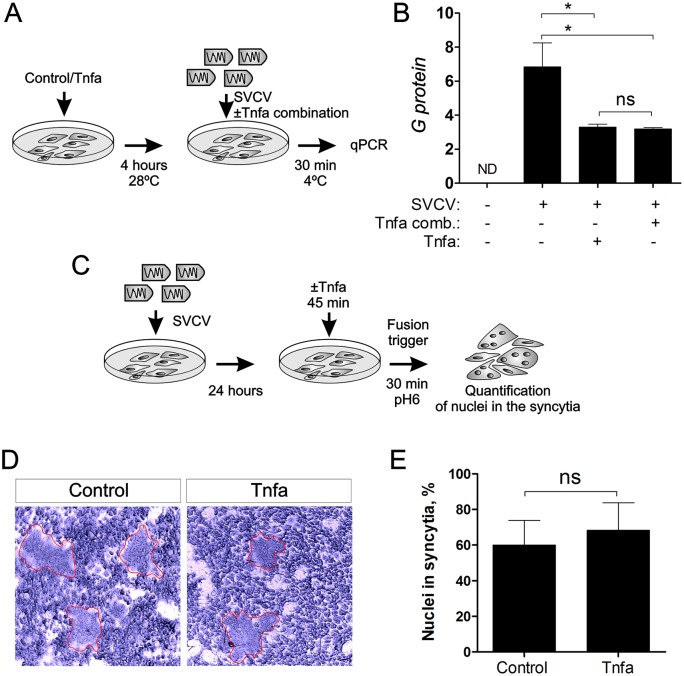
Fig 3. SVCV adhesion or syncytia production is not affected by Tnfa.
(A) Workflow of the experimental design followed in (B). Briefly, recombinant zebrafish Tnfa was added to ZF4 cells growing in monolayer at 80% of confluence and incubated for 4 hours. Subsequently, the medium was washed out and new medium containing SVCV ± Tnfa (Tnfa combination) was added for 30 minutes at 4°C to allow virus adhesion but not virus internalization and replication. After 30 minutes, the cells were washed and harvested for qPCR analysis of the G protein gene for quantitation of the adhered virus. (B) qPCR analysis of the G protein encoding gene. Bars represent mean ± S.E.M. of triplicate readings from one sample and the data are representative of two independent experiments. *p<0.1. ns, non significant. Tnfa comb., Tnfa added in combination to the SVCV. (C) Workflow representing the experimental design for (D) and (E). ZF4 were incubated with SVCV for 24 hours. The medium containing virus was washed out and new medium containing Tnfa was added for 45 minutes. The fusion process was triggered by decreasing the pH to 6 for 30 minutes, and the nuclei in the syncytia were quantitated (D, E). Red lines denote syncytia.