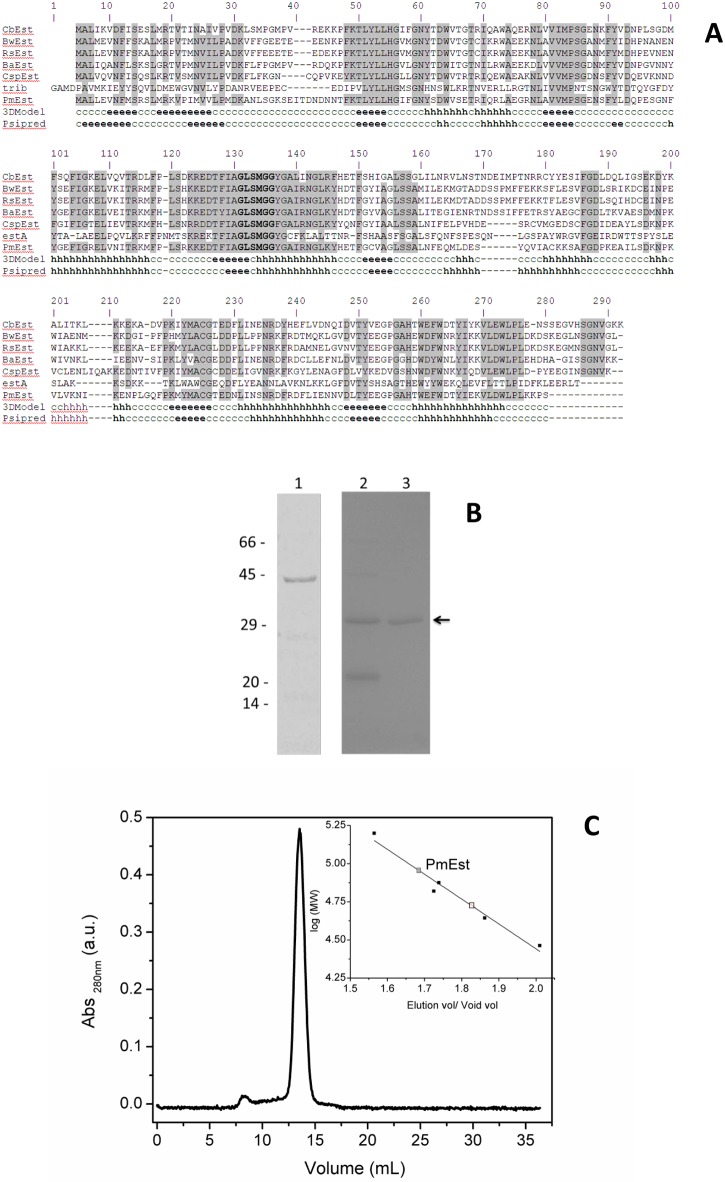
Fig 1. Characterization of the PmEst enzyme.
a) Sequence alignment between PmEst and esterases from other organisms. Conserved residues among esterases are highlighted in gray, and residues from esterase domains (GxSxGG) are in bold font. Secondary structure prediction of PmEst using Psipred and based on the 3-D homology model. Secondary structure elements are labeled as h, helix; e, extended β-strand; c, disordered/other. CbEst, acetyl esterase from Clostridiaceae bacterium GM1; BwEst acetyl esterase from Blautia wexlerae; RsEst, esterase from Ruminococcus sp. From JC304; BaEst, acetyl esterase from Bacillus akibai; CspEst, esterase from Coprobacillus sp.D7; and estA, esterase (PDB ID: 2UZ0, chain B) from Streptococcus pneumoniae. b) SDS-PAGE showing: 1) SUMO-PmEst fusion protein after affinity chromatography on Ni-NTA; 2) fusion protein after digestion with SUMO protease; 3) purified PmEst after rechromatography on Ni-NTA column; on the left, molecular weight markers in kDa; black arrow is indicating PmEst protein; c) Size exclusion chromatography on Superdex 200 10/300 GL column (13 μm average particle size) with MW calibration curve inset: carbonic anhydrase (29 kDa), ovalbumin (44 kDa), bovine serum albumin (66 kDa), conalbumin (75 kDa), and aldolase (158 kDa).