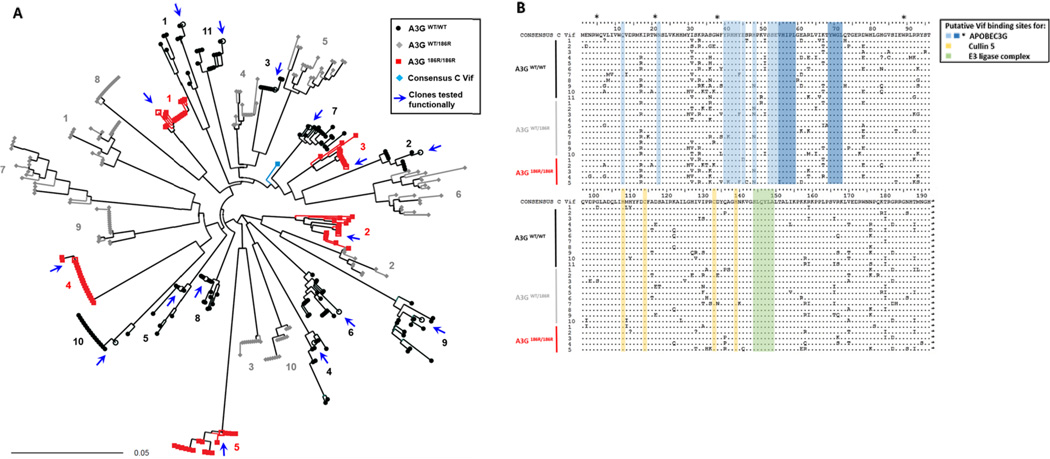
Figure 1. Sequence analysis of patient-derived HIV-1 Vif sequences.
(A) Neighbour joining phylogenetic tree of 392 full length HIV-1 vif clonal sequences shows HIV-1 vif clonal sequences from each of 26 participants forming independent clusters. The patient’s A3G genotype from which Vif clones were derived are represented by the indicated symbols and colours. Vif clones that were functionally tested are represented by open symbols and arrows. Patient samples were assigned numbers that correspond in later figures. (B) Alignment of Vif amino acid consensus sequences of 26 study samples. Sequences are compared to a consensus subtype C reference sequence (obtained from the Los Alamos National Laboratory HIV database (http://www.hiv.lanl.gov). Protein domains putatively involved in interactions that lead to proteasomal degradation of A3G are indicated; blue indicates I9, N22, E45 and N48, YRHHY (40–44), amino acids 52 to 72 including the highlighted VHIPLx4-5Lx2YWGI motif which are important for binding to A3G. * indicates tryptophan residues important for A3G binding; yellow indicates the HCCH motif important for binding to Cullin 5 and green shows SLQYLA motif important for recruitment of ubiquitin-ligase (E3) complex containing elongin –B and –C, cullin-5 and Rbx.