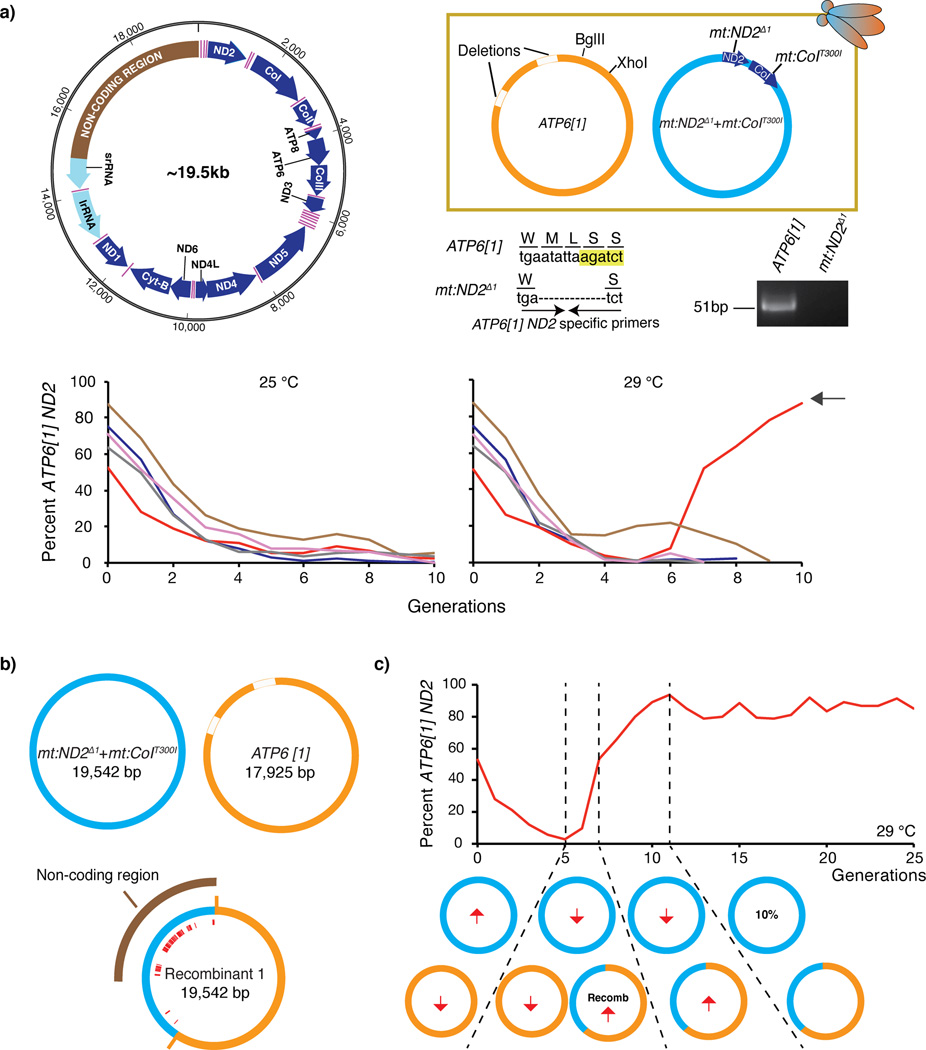
Figure 1.
Selection based on selfish drive in a heteroplasmic line containing the ATP6[1] genome and the temperature sensitive double-mutant: mt:ND2del1 + mt:CoIT300I. (a) Decline of the ATP6[1] genome when co-existing with mt:ND2del1 + mt:CoIT300I. A schematic (upper left) of D. melanogaster mitochondrial genome with protein coding genes (blue), rDNA loci (light cyan), tRNAs (pink) and the non-coding region (brown). Key features distinguishing the ATP6[1] and temperature sensitive genomes are indicated (upper right panel). A PCR primer set that selectively amplifies the intact ND2 locus of the ATP6[1] genome is indicated (BglII site, yellow highlight). The relative abundance of the ATP6[1] genome as assessed by qPCR for five lines maintained at 25 °C and 29 °C for multiple generations. After the ATP6[1] abundance fell to a low level (illustrated), the flies at 29 °C started to die (not shown), but in one line a few survivors expanded and showed an increasing abundance in a genome with the ATP6[1] ND2 region (red line, black arrow). (b) The map of the recombinant genome sequenced by PacBio SMRT technology. Red lines indicate the distribution of SNPs characteristic of mt:ND2del1 + mt:CoIT300I genome that are present in the recombinant. The ATP6[1] genome also lacks ~1.6 kb of the non-coding region17. (c) The transmission of the recombinant genome was favored when paired with the temperature sensitive genome. The directional arrows indicate how the abundance of a particular genotype was increasing or decreasing at any given generation.