Abstract
Background
Previous studies suggest that infection with non-R5-tropic subtype B HIV-1, compared to R5, is associated with a more rapid decline in CD4 count, but does not affect PI/(N)NRTI therapy outcome. Here, we explored clinical correlates associated with viral tropism in subtype A1 and D infections.
Methods
HIV-1 subtype A1 (n=196) and D (n=143) pre-therapy plasma samples and up to 7.5 years of post-therapy virologic and CD4 data were collected from a cross-sectional cohort in Mbarara, Uganda. Tropism and subtype were inferred using env V3 (geno2pheno) and gp41 (RIP) Sanger sequences. For each subtype, R5 infection was compared with non-R5 in terms of: Pre-therapy viral load and CD4 count (Mann-Whitney tests), and therapy outcomes including time to virologic suppression, post-suppression virologic rebound, CD4 decline and CD4 recovery (Log-rank tests).
Results
94% of all patients in this study achieved virologic suppression within median 3 months post-therapy. In both subtypes, non-R5 infection was associated with lower pre-therapy CD4 count (non-R5 versus R5; A: median 57 versus 147 cells/µL p=0.005; D: 80 versus 128 cells/µL p=0.006). Multivariable linear regression confirmed that tropism, not subtype nor the interaction between subtype and tropism, was a significant predictor of pre-therapy CD4 count (p<0.0001). None of pre-therapy viral load, time to virologic suppression, virologic rebound, CD4 decline nor CD4 recovery was significantly different (all p>0.09).
Conclusion
Regardless of HIV-1 subtype or tropism, the majority of patients in this Ugandan cohort responded to therapy, even though non-R5 infection was associated with lower pre-therapy CD4 count.
Keywords: Uganda, Africa, subtype A1, subtype D, non-B tropism, virologic outcome, consequence, HIV-1, clinical outcome
Introduction
HIV is genetically diverse. Subtype B HIV-1 is the most prevalent strain in Europe and North America; however, infection by this subtype accounts for just above 12% of the global epidemic [1]. Treatment-naïve individuals infected with different HIV-1 subtypes has been shown to experience varying rates of disease progression, with subtype C and D described as being more aggressive with a faster disease progression [2,3]. Another study showed that subtype D infected individuals are more likely to fail antiretroviral therapy with drug resistance mutations than those infected with subtype A [4].
Another facet of HIV genetic diversity that may impact virulence is viral tropism. HIV can be classified by its co-receptor usage: “R5-tropic HIV” infects host cells that express cell surface CCR5 receptors, whereas strains that can use alternative co-receptors including CXCR4 are collectively termed “non-R5-tropic HIV” [5,6]. Detection of non-R5-tropic HIV virus is associated with lower CD4 counts [7,8], a more advanced disease stage and a faster disease progression, but there is no clear impact of co-receptor usage on antiretroviral therapy response [9]. In a predominantly subtype B chronically infected cohort, non-R5-tropic HIV-1 was associated with a poorer pre-therapy baseline viral load and CD4 count, but tropism was not associated with therapy outcomes examined including time to virologic suppression, time to virologic rebound, time to CD4 decline and time to nonaccidental death [10]. It has not been reported whether the same trends could be observed in other HIV-1 subtypes.
The relationship between HIV-1 subtype, tropism and their associations with clinical correlates is also poorly understood. Both subtype D and non-R5 are independently associated with a faster disease progression [2,3,7,8]. Numerous studies have shown that subtype D viruses are more likely to be non-R5-tropic [7,8,11–14], and non-R5-tropic subtype D infections in Ugandan babies is linked to higher mortality [12]. These observations lead to the question of whether an infection that is both subtype D and non-R5 would lead to synergistic increase in disease progression. Little evidence exists to evaluate this hypothesis.
The objective of this study is to explore clinical correlates associated with viral tropism in subtype A1 and D HIV-1 infections. We hypothesize that in a Ugandan cohort in which subtype A1 and D HIV-1 co-circulate, having non-R5-tropic HIV-1 would be associated with a poorer pre-therapy clinical profiles, but would not differ from R5-infections in therapy outcomes. HIV-1 gp41 and V3 sequences were used to infer viral subtype and genotypic tropism respectively, which were then used to test for statistical associations against pre- and post-treatment clinical correlates. We also compared between the subtypes and explored relationships between subtype, tropism and clinical correlates.
Methods
Cohort Description
The Uganda AIDS Rural Treatment Outcomes (UARTO study) is a cross-sectional cohort of 500 initially treatment-naïve HIV-1 infected subjects followed primarily at the Immune Suppression Syndrome (ISS) Clinic in Mbarara, Uganda, a rural community 4.5 hours by automobile from the capital city of Kampala [15–17]. Participants were enrolled just prior to the start of antiretroviral regimen between June 27, 2005 and April 8, 2010, and were longitudinally followed approximately every three months to receive HIV RNA and CD4 count monitoring for up to 7.5 years until January 11, 2013 or when they were lost to follow-up. Infections were predominantly subtype A1 (49%) and D (43%) [18]. All 500 participants were included in this study. Overall adherence for the cohort was 86% [19]. Mortality data, opportunistic infections, treatment side-effects and reasons for loss to follow up were unavailable. The study was approved by the Mbarara University of Science and Technology Human Subjects Committee (14/01-03), the Uganda Council of Science and Technology (HS 938), Partners Healthcare Human Subjects Committee (2011P000522), the University of British Columbia/ Providence Health Care Research Ethics Board (H11-01642), and the University of California Human Research Subjects Committee (10-03457).
PCR and Sanger Sequencing
Total nucleic acid was extracted from 500 µL of plasma samples using NucliSENS easyMag (bioMérieux). Reverse transcription and cDNA synthesis was performed for HIV-1 env V3 loop, protease/reverse transcriptase and gp41 independently with SuperScript® III One-Step RT-PCR System (Invitrogen) followed by “nested” second-round PCR (Supplementary Tables 1 and 3) [modified based on 20,21]. Bulk sequencing was performed on ABI 3730 DNA Sequencer using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) (Supplementary Table 2). Resulting chromatograms were aligned against HXB2 and base-called using in-house software RECall [22].
Definitions of viral tropism and subtype
Viral tropism was inferred using geno2pheno (g2p) (http://coreceptor.geno2pheno.org/) [23], a bioinformatics tool that compared user input HIV-1 env V3-loop nucleic acid sequences to known V3 sequences that had matching phenotypic tropism data. g2p gave one false positive rate (fpr) value between 0–100% per input V3 sequence, representing its likelihood to be non-R5-tropic. In this study, a virus was inferred R5-tropic if g2p fpr was <5.75%, and non-R5-tropic if g2p fpr was >=5.75%. This cutoff value was chosen for our primary analysis to infer tropism because it has been shown to predict non-B HIV-1 phenotypic tropism results with similar sensitivity and specificity compared with subtype B viruses [24]. HIV-1 subtype was inferred using HIV-1 env gp41 sequences and RIP 3.0 (http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html) with a window size of 400 and confidence interval of 95%. V3 loop sequence alone (~105 nucleotides) was too short for RIP 3.0; therefore, gp41 (~1035 nucleotides) was chosen for subtype prediction because of its close proximity to V3 in the genome and its location in env. All intra-gene recombinants were excluded. Other definitions of viral tropism (g2p fpr cutoff 5%, 10%, 15%, 20% and PhenoSeq [25]) and subtype (RIP using protease/reverse transcriptase, RIP with reduced window size of 100 and REGA http://www.bioafrica.net/rega-genotype/html/subtypinghiv.html) were examined as part of the sensitivity analysis.
Definitions of Therapy Outcomes
Virologic suppression was defined as <400 copies HIV RNA/mL to reflect the viral load detection limit during the initial years of the follow-up period. Four therapy outcomes were examined: (i) Time to virologic suppression (first of two consecutive viral load <400 copies HIV RNA/mL), (ii) Time to virologic rebound (first of two consecutive viral load ≥400 copies HIV RNA/mL post-suppression), (iii) Time to any CD4 decline below baseline, and (iv) Time to CD4 recovery (first of two consecutive post-therapy CD4 count increase of >200 cells/µL from pre-therapy count, or first of two consecutive post-therapy CD4 count >350 cells/µL). Other definitions of virologic suppression (50 and 500 copies HIV RNA/mL) and CD4 recovery (time to first of two consecutive post-therapy CD4 count above baseline) were examined as part of the sensitivity analysis.
Statistical Methods
All statistical analyses were performed using R and/or GraphPad Prism 5.0. Two-tailed Mann-Whitney tests were used to compare pre-therapy viral load, pre-therapy CD4 count, g2p %fpr, and time to loss to follow-up. Log-rank tests were used to compare Kaplan-Meier curves of time to virologic suppression, post-suppression virologic rebound, CD4 decline and CD4 recovery. Two-tailed Fisher's exact test was used to compare the prevalence of dichotomized non-R5 and R5 in subtype A1 and D.
A multivariable linear regression was used to explore the effect of viral subtype, viral tropism, gender, age, and baseline viral load on baseline CD4 count. In this analysis, the square-root of baseline CD4 count was used to obtain normality as previously described [16,17]; normality of the transformed data was confirmed by Q-Q plot. Confidence intervals (CI) at 95% for each coefficient are reported in the result section as CI: [lower, upper].
Statistical significance was defined as p<0.05. Note, due to the large number of statistical tests performed in the sensitivity analyses, Bonferroni correction should arguably be used to adjust the threshold for statistical significance to p = 0.05/470 = 0.0001.
Results
Pre-therapy baseline characteristics and overall therapy outcomes
Based on our definitions of viral subtype (RIP, gp41) and tropism (g2p 5.75%fpr cutoff, env V3), 339 of the 500 UARTO study participants were infected with subtype A1 or D HIV-1 and had an inferred V3 tropism result: 231 (68%) were female, and 196 (58%) were infected with subtype A1. Median age was 35 years (Q1–Q3 30–39), baseline viral load 1×105 copies HIV RNA/mL (Q1–Q3 4×104–4×105), and baseline CD4 count 129 cells/µL (Q1–Q3 70–202). Initial regimens were primarily NVP- (86%) and EFV-based (12%) in combination with lamivudine (3TC) and zidovudine (AZT) as backbones.
Longitudinal viral load and CD4 data were available for n=315 of these patients: 94% achieved virologic suppression (defined as first of two consecutive viral load <400 copies HIV RNA/mL) within a median of three months post-therapy, 10% of those who achieved suppression experienced subsequent virologic rebound (defined as first of two consecutive viral load ≥400 copies HIV RNA/mL post-suppression) over a median of 1.4 years post-suppression, and 72% achieved CD4 recovery (first of two consecutive post-therapy CD4 count increase of >200 cells/µL from pre-therapy count, or first of two consecutive post-therapy CD4 count >350 cells/µL) in a median of 2.2 years.
Primary Analysis: Clinical correlates of R5 versus non-R5 infections
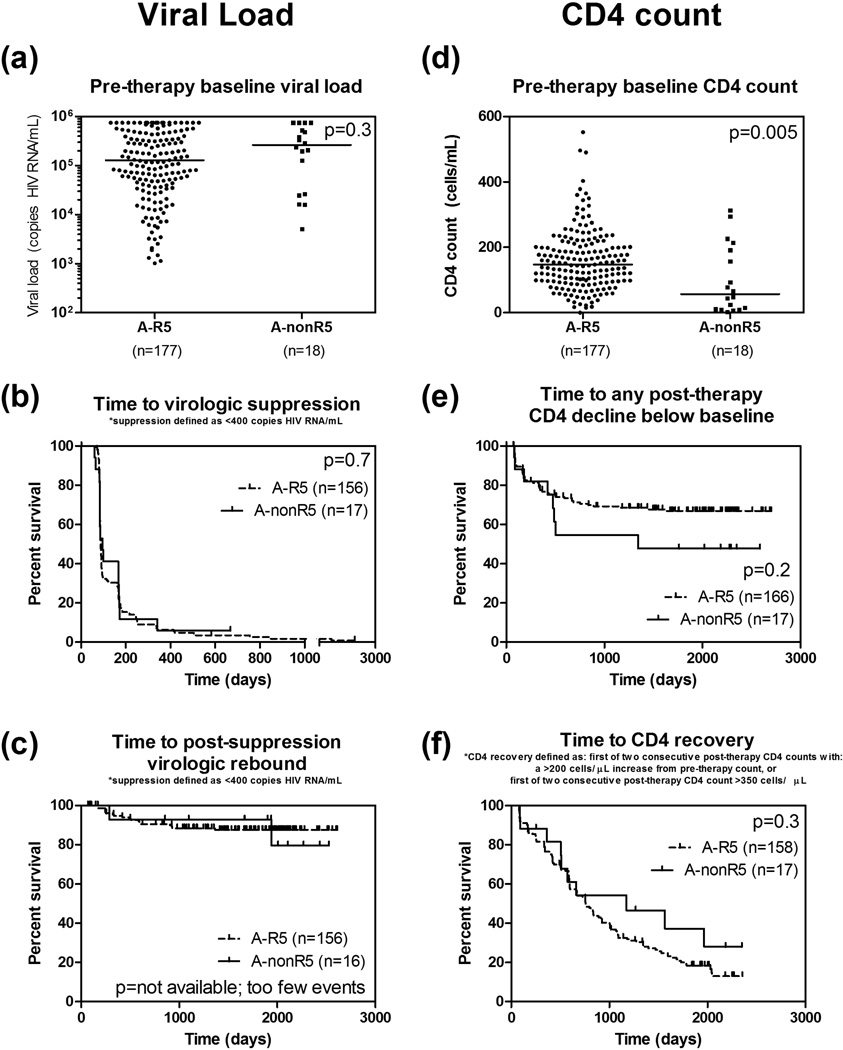
In subtype A1 infections, non-R5 (n=178) had significantly lower baseline CD4 count than R5 (n=18) (median 57 versus 147 cells/µL, p=0.005). No other clinical correlates or outcomes tested was significantly different including baseline viral load (p=0.3), time to virologic suppression (p=0.7), time to CD4 decline below baseline (p=0.2) and time to CD4 recovery (p=0.3) (Figure 1).
Figure 1.
In subtype A1 HIV-1 infections, detection of non-R5 viruses was significantly associated with lower pre-therapy baseline CD4 count (d), but not with pre-therapy baseline viral load (a), time to virologic suppression (b), time to virologic rebound (c), time to CD4 decline below baseline (e), and time to CD4 recovery (f). Horizontal lines indicate median.
In subtype D infections, the same pattern was observed: Non-R5 had lower baseline CD4 count than R5 (median 80 versus 128 cells/µL, p=0.006). No other clinical correlates or outcomes tested was statistically different including baseline viral load (p=0.4), time to virologic suppression (p=0.3), time to CD4 decline below baseline (p=0.5) and time to CD4 recovery (p=0.09) (Figure 2). Note that only two patients with non-R5 subtype A1 infections, and only one patient with non-R5 subtype D infection, experienced post-therapy virologic rebound over the course of maximum 7.5 years of longitudinal follow-up. As a result, statistical tests to compare time to virologic rebound between groups could not be performed due to the lack of statistical power.
Figure 2.
Similar to subtype A1, in subtype D HIV-1 infections, detection of non-R5 viruses was significantly associated with lower pre-therapy baseline CD4 count (d), but not with pre-therapy baseline viral load (a), time to virologic suppression (b), time to virologic rebound (c), time to CD4 decline below baseline (e), and time to CD4 recovery (f). Horizontal lines indicate median.
To examine potential bias introduced by differences in loss to follow-up, we compared time to loss to follow-up between groups in all of the above “time to event” analyses. We observed no significant differences between groups (all p>0.1).
Secondary Analysis: Subtype A1 versus D
In addition to our primary analysis, we also observed that patients with subtype A1 viruses had significantly lower prevalence of non-R5 HIV-1 than those with subtype D (9% versus 41%, p<10−11); this was in accordance with their striking difference in g2p %fpr distribution (median 64% fpr versus 9% fpr, p<10−14) (Figure 3). Furthermore, we observed that patients with subtype A1 infections had significantly higher baseline CD4 count than D (median 144 versus 114 cells/µL, p=0.007); no other clinical correlates or outcomes tested were statistically significant (all p>0.09). Median time to loss to follow-up was not different between groups (p=0.7). Mortality data and reasons for loss to follow-up were unavailable.
Figure 3.
Geno2pheno (g2p) % false positive rate (fpr) values derived from HIV-1 subtype A1 env V3 sequences were significantly higher than subtype D. Horizontal lines indicate median.
Based on the above observations, we hypothesized that this lower pre-therapy baseline CD4 count observed in subtype D infections, compared to subtype A1, was driven by viral tropism and not by viral subtype. Indeed, baseline CD4 count in subtype A1 R5 and subtype D R5 HIV-1 infections were not statistically different (median 147 versus 128 cells/µL, p=0.2). The same was true for subtype A1 versus subtype D non-R5 HIV-1 infections (median 57 versus 80 cells/µL, p=0.6). In a multivariable linear regression model with baseline CD4 count as the response variable, tropism was found to be a significant predictor (CI: [−6×10−6, −3×10−6], p<10−5) while controlling for gender (CI: [−0.03, 1.9], p=0.06), age (CI: [−0.01, 0.1], p=0.1), subtype (CI: [−1.7, 0.1], p=0.1) and baseline viral load (CI: [−3.7, −1.5], p<10−6). Subtype and the interaction between subtype and tropism were not found to be significant predictors of baseline CD4 count.
Sensitivity Analyses
To evaluate the effect of using another part of the HIV genome for subtype prediction, protease/reverse transcriptase was chosen because it is arguably the most commonly sequenced part of the HIV genome globally due to genotypic resistance testing [26]. Among all pre-therapy protease/reverse transcriptase sequences (n=454), 44% were subtype A1, and 37% were D. Absolute concordance between subtype predictions within a patient using protease/reverse transcriptase versus gp41 to infer subtype was 66%, indicating a high prevalence of recombination in this cohort. Using this alternative subtype definition, a similar pattern was observed in both subtypes: Non-R5 had lower baseline CD4 count than R5 infections (A: p=0.02; D: p=0.002); no other clinical correlates or outcomes tested were statistically different (all p>0.08).
Sensitivity analyses were also performed to examine whether a different RIP window size (100), a different HIV-1 subtyping tool (REGA), a different and subtype-specific algorithm to infer viral tropism (PhenoSeq [25]), different g2p fpr % cutoffs (5%, 10%, 15%, 20%), different definitions of virologic suppression (50 and 500 copies HIV RNA/mL), or a different measure of CD4 recovery (time to first of two consecutive post-therapy CD4 above baseline) would yield different observations. In summary, all yielded trends similar to our primary analysis regardless of HIV gene used to infer subtype, g2p fpr % cutoff values, and the definition of virologic suppression or CD4 recovery (results not shown).
Discussion
In summary, we found that in both subtype A1 and D infections, non-R5-tropic HIV-1 was associated with lower pre-therapy baseline CD4 count, but not with pre-therapy baseline viral load, nor with any of the post-therapy outcomes examined. Furthermore, we identified viral tropism as inferred from HIV-1 V3 sequence, but not subtype, as the apparent driver for the observed association between both non-R5 tropism and subtype D with a lower pre-therapy baseline CD4 count.
Neither subtype nor tropism affected PI/(N)NRTI therapy outcomes in subtype A1 or D, R5 or non-R5 infections for an extended period of time. Overall, 94% of study participants achieved virologic suppression within three months post-therapy. We did not observe sufficient cases of post-therapy viral load rebound, to the extent that it was impossible to perform meaningful statistical tests even after 7.5 years of follow-up time. This implies that even if non-R5 tropism had led to an increased frequency of viral load rebound, the magnitude of its effect must be small. This finding is similar to previous findings in subtype B patients [10]. Together, our observations suggest that HIV care architecture was highly effective at least in this particular rural Ugandan cohort setting; specifically, the patients received frequent, 3–6 months-apart clinical monitoring and adherence was at a median of 86% [19]. However, it should also be noted that our conclusion is limited by the lack of mortality data; individuals who did not experience virologic rebound could either be lost to follow-up due to non-AIDS related reasons, or could have died without a chance for us to observe two consecutive rise in viral load. To shed more light on the effect of this limitation, we compared time to loss to follow-up between the groups, but did not observe significant differences.
Another major question addressed by our study is the relationship between subtype, tropism and pre-therapy CD4 count. Previous studies tend to focus on only two of the three factors at a time: In terms of the relationship between HIV-1 subtype and pre-therapy CD4 count, our observation agrees with previous reports that pre-therapy subtype D infections are “more aggressive” than subtype A1 by being associated with a lower baseline CD4 count [2,3]. In terms of the relationship between HIV-1 subtype and viral tropism, our observation agrees with previous reports that subtype D is associated with a higher prevalence of non-R5-tropic viruses [7,8,11–14]. In terms of the relationship between viral tropism and pre-therapy CD4 count, our observation agrees with previous reports that non-R5 infections are associated with lower CD4 count [10,27,28].
Unlike any of the aforementioned studies [2,3,7,8,10–14,27,28] that focused on only two factors at a time, we statistically examined the relationship between all three factors (subtype, tropism and pre-therapy CD4 count) by comparing baseline correlates and clinical outcomes between R5 infections in subtype A1 against D, as well as non-R5 infection in subtype A1 against D, and found no significant differences between groups. In other words, an infection that was both subtype D and non-R5 did not show a synergistic and negative effect at baseline when compared to subtype A non-R5 infections, suggesting that viral tropism, not subtype, drove the previously observed association between both non-R5 tropism and subtype D with a lower pre-therapy baseline CD4 count. Our multivariable linear regression model confirmed that the association between subtype D infections and a poorer pre-therapy baseline CD4 count was attributed to tropism, but not to viral subtype, after adjusting for age, gender and baseline viral load. A similar trend was reported in HIV-1 CRF01_AE compared to CRF07_BC and subtype B infections [29].
This conclusion that pre-therapy baseline CD4 count was associated with tropism not subtype was limited by the cross-sectional nature of this study and the lack of time-since-infection data, a factor strongly associated with CD4 depletion over time (reviewed in [30]). We cannot rule out that patients infected with non-R5 viruses were simply infected for a longer period of time and were at a more advanced disease stage than those infected with R5 viruses. The casual relationship between this commonly observed association between non-R5 viruses and advanced disease stage is still poorly understood [9].
Another study limitation was the methods used for tropism and subtype prediction. It is possible that the reliability of genotypic tropism prediction algorithms like g2p is itself dependent on viral subtypes. We have previously reported that g2p with 5.75% fpr cutoff demonstrated lower sensitivity in subtype A1 (44.4%) and a lower specificity (68.2%) in subtype D viruses compared to subtype B (sensitivity 65.6%, specificity 90.5%) against Trofile phenotypic tropism assay, and other algorithms such as PSSM showed similar performance [24]. In the same study [24], we also showed that adjusting the algorithms’ cutoff value may further optimize sensitivity and specificity for each subtype. Hence in the current study, we performed sensitivity analyses by redefining tropism with g2p cutoffs at 5%, 10%, 15% and 20%, and with a subtype-specific tropism inference tool PhenoSeq [25]; overall, all yielded trends similar to our primary analysis with g2p cutoff at 5.75%.
In terms of subtype prediction, we have chosen RIP 3.0 with window size 400 among the myriad of subtype inferring tools available; these subtype prediction tools do not generate perfectly concordant results [31]. However, reanalyses using a more relaxed RIP window size of 100 as well as another HIV-1 subtyping tool REGA did not change our conclusions. Furthermore, we have demonstrated that using different parts of the HIV genome to infer subtype in the UARTO cohort introduced discordance due to a high prevalence of intra-genome recombinants between subtypes. This problem was further explored in another study [32]. In this study, all intra-gene recombinants were excluded to avoid potential bias.
In summary, our study showed that pre-therapy baseline CD4 count was associated with viral tropism, not subtype. We also showed that an overall 94% of study participants in this rural Ugandan cohort achieved virologic suppression within three month post-therapy, regardless of viral subtype or tropism, an indication that the HIV care architecture in this rural Ugandan village was highly effective.
Supplementary Material
Acknowledgments
Source of Funding: PRH has received consulting fees from ViiV/Pfizer and Quest; holds stock in Merck, Gilead and Illumina. This work was funded by the Canadian Institutes of Health Research (CIHR), National Institutes of Health (NIH) Centers for AIDS Research (CFAR) Program, National Institute of Mental Health R01MH054907, P30AI027763, U01CA066529 and National Institute on Alcohol Abuse and Alcoholism R21AA014784.
We thank all UARTO study participants for making this work possible. We also thank Dr Kieran Cashin for his help with PhenoSeq viral tropism inferences.
Footnotes
Presented in part at: The 54th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC 2014), Washington DC, US (Abstract H-1633)
Conflicts of Interest: For the remaining authors no conflicts of interest were declared.
Authors contribution: The work presented here was carried out in collaboration between all authors. The study was conceptualized and designed by GQL and PRH. Plasma samples, baseline and follow-up data were collected and managed by ARM, DRB, JNM, PWH, and YB. HIV-1 genotyping laboratory work was done by CL and GQL. Results were analyzed by EC, GQL and VDL. GQL wrote the manuscript; all authors contributed to, seen, and approved the manuscript.
References
- 1.Geretti AM. HIV-1 subtypes: epidemiology and significance for HIV management. Curr Opin Infect Dis. 2006;19:1–7. doi: 10.1097/01.qco.0000200293.45532.68. [DOI] [PubMed] [Google Scholar]
- 2.Santoro MM, Perno CF. HIV-1 Genetic Variability and Clinical Implications. ISRN Microbiol. 2013;2013:481314. doi: 10.1155/2013/481314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pant Pai N, Shivkumar S, Cajas JM. Does genetic diversity of HIV-1 non-B subtypes differentially impact disease progression in treatment-naive HIV-1-infected individuals? A systematic review of evidence: 1996–2010. J Acquir Immune Defic Syndr. 2012;59:382–388. doi: 10.1097/QAI.0b013e31824a0628. [DOI] [PubMed] [Google Scholar]
- 4.Kyeyune F, Nankya I, Metha S, Akao J, Ndashimye E, Tebit DM, et al. Treatment failure and drug resistance is more frequent in HIV-1 subtype D versus subtype A-infected Ugandans over a 10-year study period. AIDS. 2013;27:1899–1909. doi: 10.1097/QAD.0b013e3283610ec7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swenson LC, Däumer M, Paredes R. Next-generation sequencing to assess HIV tropism. Curr Opin HIV AIDS. 2012;7:478–485. doi: 10.1097/COH.0b013e328356e9da. [DOI] [PubMed] [Google Scholar]
- 6.Lee GQ, Cheung PK, Swenson LC, Harrigan PR. Assessment of HIV-1 tropism using genotypic approaches. Hot Top HIV Other Retroviruses. 2012;2:7–13. [Google Scholar]
- 7.Ataher Q, Portsmouth S, Napolitano La, Eng S, Greenacre A, Kambugu A, et al. The epidemiology and clinical correlates of HIV-1 co-receptor tropism in non-subtype B infections from India, Uganda and South Africa. J Int AIDS Soc. 2012;15:2. doi: 10.1186/1758-2652-15-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaleebu P, Nankya IL, Yirrell DL, Shafer LA, Kyosiimire-Lugemwa J, Lule DB, et al. Relation between chemokine receptor use, disease stage, and HIV-1 subtypes A and D: results from a rural Ugandan cohort. J Acquir Immune Defic Syndr. 2007;45:28–33. doi: 10.1097/QAI.0b013e3180385aa0. [DOI] [PubMed] [Google Scholar]
- 9.Verhofstede C, Nijhuis M, Vandekerckhove L. Correlation of coreceptor usage and disease progression. Curr Opin HIV AIDS. 2012;7:432–439. doi: 10.1097/COH.0b013e328356f6f2. [DOI] [PubMed] [Google Scholar]
- 10.Brumme ZL, Goodrich J, Mayer HB, Brumme CJ, Henrick BM, Wynhoven B, et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis. 2005;192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 11.Church JD, Huang W, Mwatha A, Toma J, Stawiski E, Donnell D, et al. HIV-1 tropism and survival in vertically infected Ugandan infants. J Infect Dis. 2008;197:1382–1388. doi: 10.1086/587492. [DOI] [PubMed] [Google Scholar]
- 12.Church JD, Huang W, Mwatha A, Musoke P, Jackson JB, Bagenda D, et al. Analysis of HIV tropism in Ugandan infants. Curr HIV Res. 2010;8:498–503. doi: 10.2174/157016210793499187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol. 2007;81:7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wambui V, Kiptoo M, Kinyua J, Odera I, Muge E, Muiruri P, et al. Predicted HIV-1 coreceptor usage among Kenya patients shows a high tendency for subtype d to be cxcr4 tropic. AIDS Res Ther. 2012;9:22. doi: 10.1186/1742-6405-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaida A, Matthews LT, Kanters S, Kabakyenga J, Muzoora C, Mocello aR, et al. Incidence and Predictors of Pregnancy among a Cohort of HIV-Positive Women Initiating Antiretroviral Therapy in Mbarara, Uganda. PLoS One. 2013;8:e63411. doi: 10.1371/journal.pone.0063411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt PW, Cao HL, Muzoora C, Ssewanyana I, Bennett J, Emenyonu N, et al. Impact of CD8+ T-cell activation on CD4+ T-cell recovery and mortality in HIV-infected Ugandans initiating antiretroviral therapy. AIDS. 2011;25:2123–2131. doi: 10.1097/QAD.0b013e32834c4ac1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byakwaga H, Boum Y, Huang Y, Muzoora C, Kembabazi A, Weiser SD, et al. The Kynurenine Pathway of Tryptophan Catabolism, CD4+ T-Cell Recovery, and Mortality Among HIV-Infected Ugandans Initiating Antiretroviral Therapy. J Infect Dis. 2014:1–9. doi: 10.1093/infdis/jiu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee GQ, Bangsberg DR, Muzoora C, Boum Y, Oyugi JH, Emenyonu N, et al. Prevalence and Virologic Consequences of Transmitted HIV-1 Drug Resistance in Uganda. AIDS Res Hum Retroviruses. 2014;30:896–906. doi: 10.1089/aid.2014.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haberer JE, Musinguzi N, Boum Y, Siedner MJ, Mocello AR, Hunt PW, et al. Duration of Antiretroviral Therapy Adherence Interruption Is Associated With Risk of Virologic Rebound as Determined by Real-Time Adherence Monitoring in Rural Uganda. 2015;02114:386–392. doi: 10.1097/QAI.0000000000000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee GQ, Dong W, Mo T, Knapp DJHF, Brumme CJ, Woods CK, et al. Limited Evolution of Inferred HIV-1 Tropism While Viremia Is Undetectable during Standard HAART Therapy. PLoS One. 2014;9:e99000. doi: 10.1371/journal.pone.0099000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brumme ZL, Dong WWY, Yip B, Wynhoven B, Hoffman NG, Swanstrom R, et al. Clinical and immunological impact of HIV envelope V3 sequence variation after starting initial triple antiretroviral therapy. AIDS. 2004;18:F1–F9. doi: 10.1097/00002030-200403050-00001. [DOI] [PubMed] [Google Scholar]
- 22.Woods CK, Brumme CJ, Liu TF, Chui CKS, Chu AL, Wynhoven B, et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012;50:1936–1942. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007;25:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 24.Lee GQ, Harrigan PR, Dong W, Poon AFY, Heera J, Demarest J, et al. Comparison of Population and 454 “Deep” Sequence Analysis for HIV Type 1 Tropism Versus the Original Trofile Assay in Non-B Subtypes. AIDS Res Hum Retroviruses. 2013;29:979–984. doi: 10.1089/aid.2012.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cashin K, Gray LR, Harvey KL, Perez-Bercoff D, Lee GQ, Sterjovski J, et al. Reliable Genotypic Tropism Tests for the Major HIV-1 Subtypes. Sci Rep. 2015;5:8543. doi: 10.1038/srep08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization. SURVEILLANCE OF HIV DRUG RESISTANCE IN ADULTS INITIATING ANTIRETROVIRAL THERAPY (PRETREATMENT HIV DRUG RESISTANCE) WHO Publ. 2014
- 27.Ribeiro RM, Hazenberg MD, Perelson AS, Davenport MP. Naive and Memory Cell Turnover as Drivers of CCR5-to-CXCR4 Tropism Switch in Human Immunodeficiency Virus Type 1 : Implications for Therapy. Society. 2006;80:802–809. doi: 10.1128/JVI.80.2.802-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunt PW, Harrigan PR, Huang W, Bates M, Williamson DW, McCune JM, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis. 2006;194:926–930. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Xue Y, Zhou L, Lin Y, Yu X, Wang X, et al. Evidence that HIV-1 CRF01-AE is associated with low CD4+T cell count and CXCR4 co-receptor usage in recently infected young men who have sex with men (MSM) in Shanghai, China. PLoS One. 2014;9:1–9. doi: 10.1371/journal.pone.0089462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okoye AA, Picker LJ. CD4 + T-cell depletion in HIV infection : mechanisms of immunological failure. Immunol Rev. 2013;254:54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan PA, Reitsma MB, Delong A, Boucek B, Nunn A, Salemi M, et al. Phylogenetic and geospatial evaluation of HIV-1 subtype diversity at the largest HIV center in Rhode Island. Infect Genet Evol. doi: 10.1016/j.meegid.2014.03.027. Published Online First: 8 April 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee GQ, Bangsberg DR, Lachowski C, Brumme CJ, Boum Y, Mocello AR, et al. Prevalence and Clinical Impacts of HIV-1 Intersubtype Recombinants in Uganda; 8th IAS Conference on HIV Pathogenesis, Treatment and Prevention (IAS 2015); Vancouver, British Columbia. 2015. pp. Abstract #A–729–0071–00276. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.