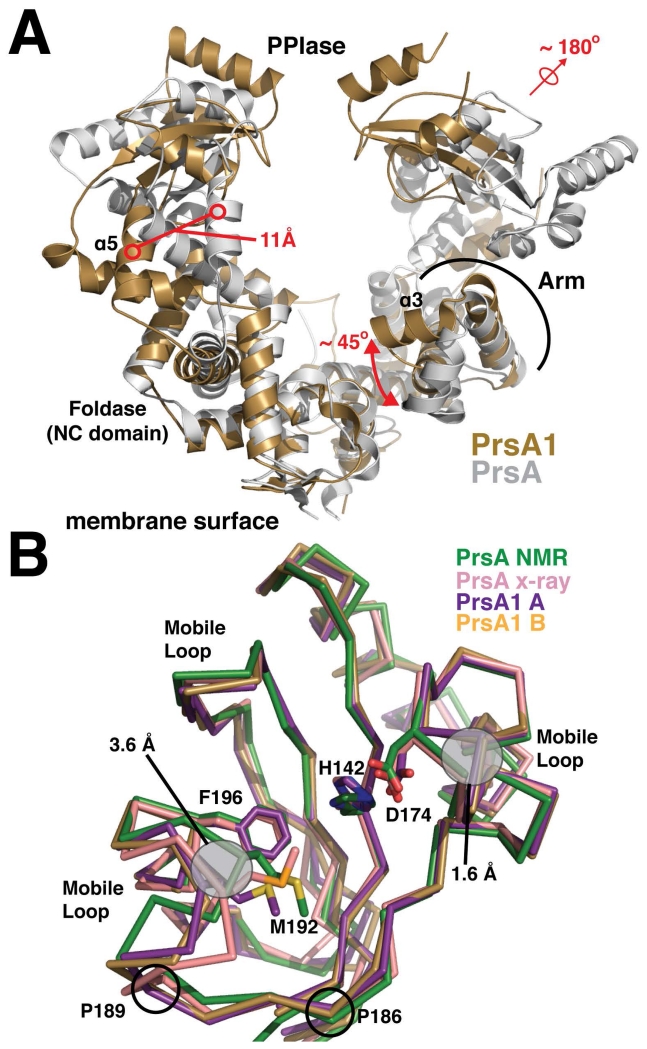
Figure 2. Structural alignment of the PrsA1 and PrsA structures.
(A) Listeria monocytogenes PrsA1 (gold) and Bacillus subtilis PrsA (PDB accession code 4WO7) (grey) are shown. The structures are aligned by an overlay of the foldase (NC) domains. Significant structural differences are highlighted in red. (B) The PPIase domains from each monomer of PrsA1 (purple, gold) are aligned with the PrsA PPIase from Bacillus subtilis (green and pink, PDB accession codes 1ZK6 and 4WO7). The side chains of several conserved catalytic residues are drawn in stick representation for comparison with conformational differences outlined in grey. The approximate regions experiencing dynamic motion as indicated from the PrsA NOE relaxation data are marked as mobile loops. The position of conserved proline residues found in both PrsA1 and PrsA2, but not in PrsA are indicated.