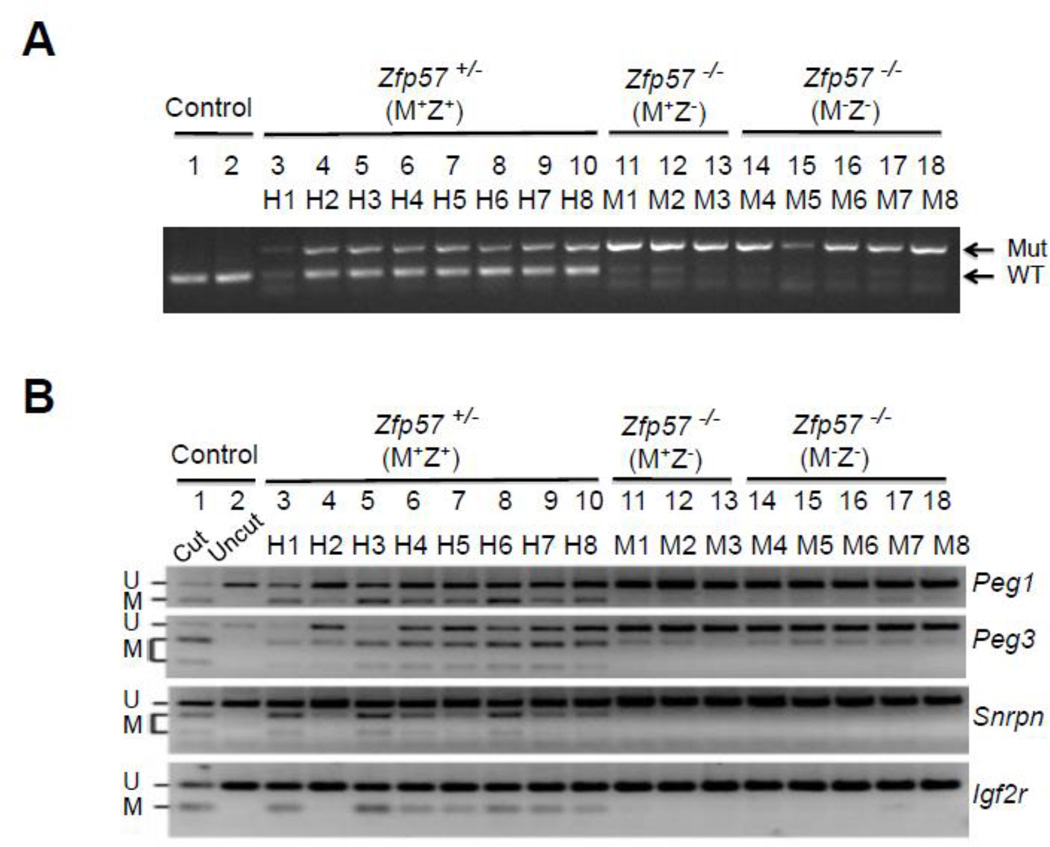
Figure 3. COBRA analysis of maternally inherited DNA methylation imprint.
A, PCR genotyping of these ES clones, similar to our previously published study (Li et al., 2008). Mut and WT, the marked gel positions for the PCR product of the mutant (Mut) and wild-type (WT) alleles of Zfp57. B, COBRA analysis was carried out for analyzing DNA methylation imprint at the maternally inherited Peg1, Peg3, Snrpn and Igf2r imprinted regions. Restriction enzyme (RE) digestion was performed for the bisulphite PCR product of all ES samples, and “Cut” (Lane 1) but not “Uncut” (Lane 2) control wild-type mouse tail DNA sample. U and M, unmethylated (U) and methylated (M) product after RE digestion, respectively. Lanes 3–10, eight Zfp57+/− (M+Z+) ES clones derived from the blastocysts generated from the cross between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice. Lanes 11–13, three Zfp57−/− (M+Z−) ES clones derived from the blastocysts generated from the cross between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice. Lanes 14–18, five Zfp57−/− (M−Z−) ES clones derived from the cross between Zfp57−/− homozygous female mice and Zfp57−/− homozygous male mice.