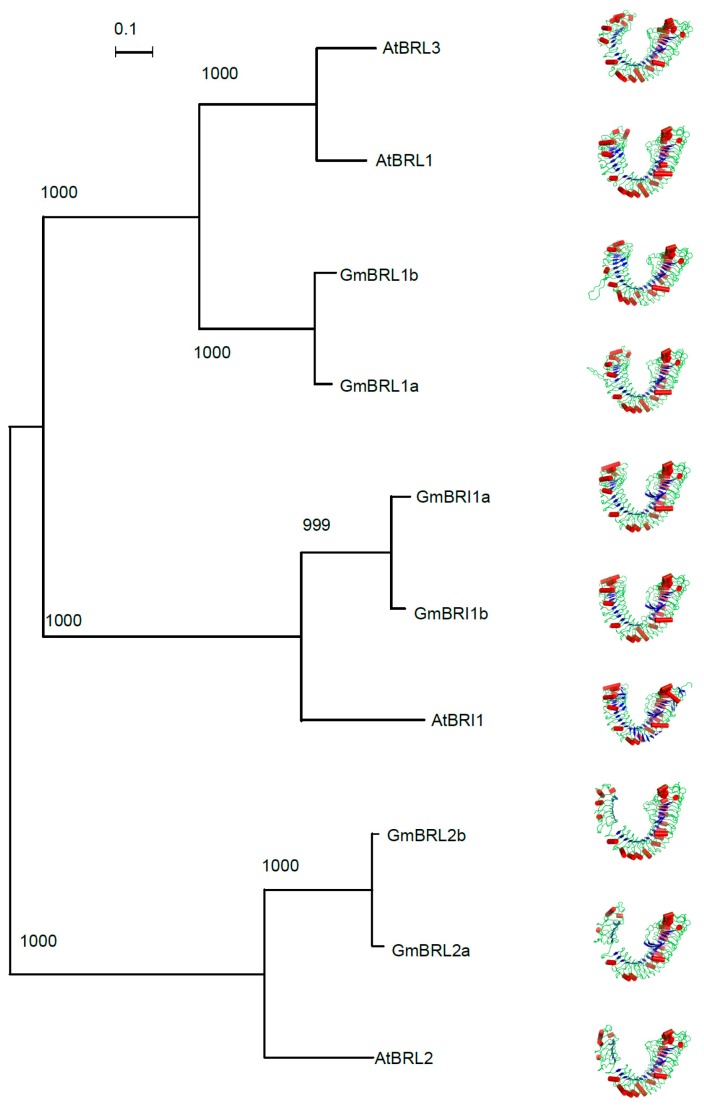
Figure 9.
Structural modeling of six soybean BR receptors. The program PhyML3.0 [58] was used to reconstruct the phylogenetic tree of BR receptors from Arabidopsis and soybean. The evolutionary lineages of four BR receptors in Arabidopsis and six BR receptors in soybean were compared. The LG model for amino acid substitutions with estimated Gamma distribution was used to reconstruct the tree and the bootstrap value was set as 1000. A total of ten BR receptors were classified into Clades I, II, and III. The numbers above each branch of the tree are the bootstrap values. Scale bar indicates 0.1 amino acid substitution over evolution. In addition, the ectodomains of ten BR receptors were modeled with MODELLER9.11 software [59] based on the template of AtBRI1, which was determined with X-ray crystallization on a 3-D level in 2011 [28]. The PDB files were processed with PyMOL v1.5 software [60].