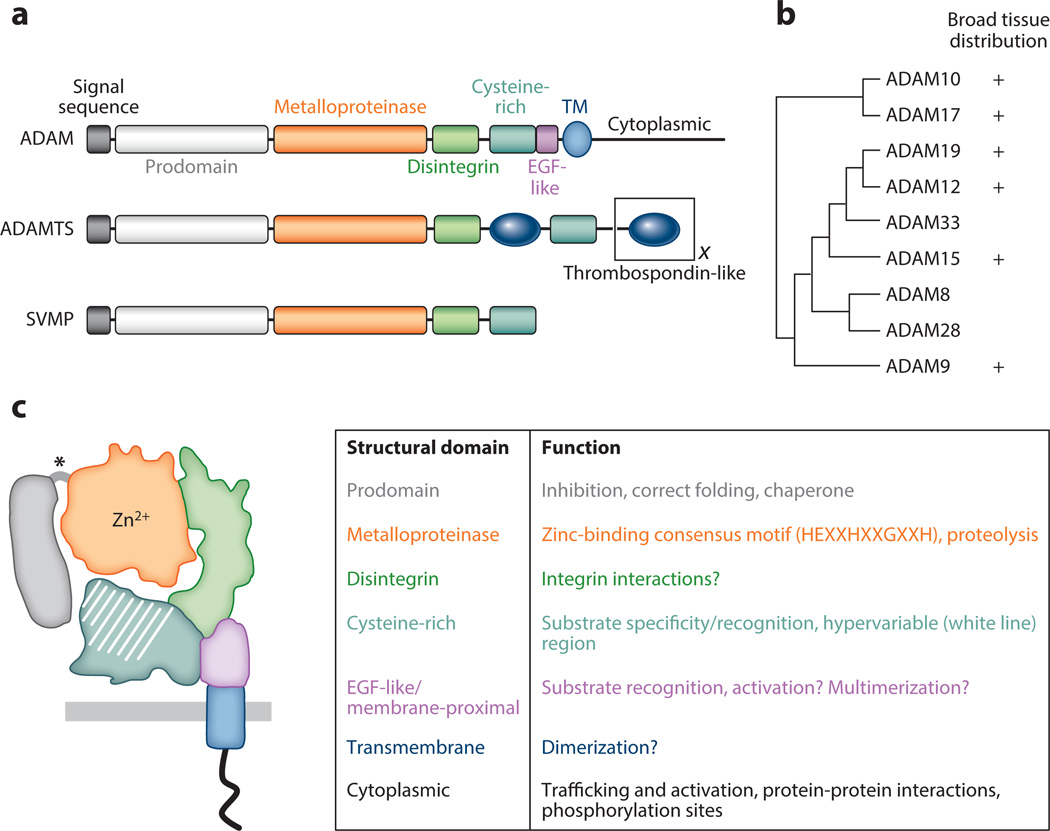
Figure 1.
Proteolytically active A disintegrin and metalloproteinases (ADAMs): structure and function. (a) ADAMs are members of the adamalysin subfamily of metzincin metalloproteinases, which also includes ADAMs containing thrombospondin motifs (ADAMTSs) and snake venom metalloproteinases (SVMPs). Different ADAMTSs contain variable numbers of thrombospondin-like motifs (represented by the black X) and other functional domains at the C terminus (not shown). Most ADAMs are synthesized as transmembrane (TM) proteins, whereas all ADAMTSs and SVMPs are secreted proteins. Another important difference between the proteins is that all SVMPs and ADAMTSs are predicted to be catalytically active, but only ~60% of ADAMs have intact metalloproteinase domains capable of proteolytic activity. (b) Phylogenetic tree built from aligned full-length amino acid sequences (left; tree adapted from References 2 and 7) and the tissue distribution of all human proteolytically active ADAMs (right). (c) ADAM domain structure and function. (Left) Color representation of structural domains: prodomain, metalloproteinase domain, disintegrin domain, cysteine-rich domain, EGF-like/proximal membrane domain, transmembrane domain, and cytoplasmic domain. Putative furin-like cleavage sites between the prodomain and metalloproteinase domain are shown by an asterisk. (Right) Summary of domain functions. Many ADAMs display broad tissue distribution, but detailed analysis of their cellular expression in different tissues is lacking.