ABSTRACT
CRISPR (Clustered Regularly-Interspaced Short Palindromic Repeats)-Cas9 (CRISPR associated protein 9) has rapidly become the most promising genome editing tool with great potential to revolutionize medicine. Through guidance of a 20 nucleotide RNA (gRNA), CRISPR-Cas9 finds and cuts target protospacer DNA precisely 3 base pairs upstream of a PAM (Protospacer Adjacent Motif). The broken DNA ends are repaired by either NHEJ (Non-Homologous End Joining) resulting in small indels, or by HDR (Homology Directed Repair) for precise gene or nucleotide replacement. Theoretically, CRISPR-Cas9 could be used to modify any genomic sequences, thereby providing a simple, easy, and cost effective means of genome wide gene editing. However, the off-target activity of CRISPR-Cas9 that cuts DNA sites with imperfect matches with gRNA have been of significant concern because clinical applications require 100% accuracy. Additionally, CRISPR-Cas9 has unpredictable efficiency among different DNA target sites and the PAM requirements greatly restrict its genome editing frequency. A large number of efforts have been made to address these impeding issues, but much more is needed to fully realize the medical potential of CRISPR-Cas9. In this article, we summarize the existing problems and current advances of the CRISPR-Cas9 technology and provide perspectives for the ultimate perfection of Cas9-mediated genome editing.
KEYWORDS: CRISPR-Cas9, efficiency, genome editing, specificity
Introduction
The gRNA-directed CRISPR-Cas9 nuclease technology has been a long awaited tool for manipulation of genomes at will. The successful application of this technology in a wide range of biological systems from yeast, worm, insect, plant, and mammals from rodent to monkey has made it the most popular genome editing technology in history.1-8 Additionally, the inactive form of CRISPR-Cas9 (dCas9) has been used in gene expression profiling and genome wide gene screening.8-10 The applicability of the CRISPR-Cas9 technology in stem cells, induced pluripotent stem cells and somatic tissues of humans forecasts its revolutionary potentials in medicine.10-16 However, the great promises of the CRISPR-Cas9 technology are paired with serious concerns about off-target activities, variations in efficacy, and limited genome coverage due to PAM restriction.17-21 The imperfection of the CRISPR-Cas9 technology is primarily due to its functional mechanism that completely depends on a short nucleotide gRNA and PAM as well as its compromising feature permitting cutting of protospacer DNAs with nucleotide mismatches to the corresponding gRNAs. The off-target activity is especially of great concern in medicine where it could produce unwanted and potentially pathologic consequences. Significant advances have been made to address these issues by crystallization of the CRISPR-Cas9-gRNA-protospacer DNA complex at different stages to illustrate its mechanistic details in target DNA binding and interrogation, which has helped facilitate the rational design of better versions of CRISPR-Cas9.22-28 Efforts on other frontlines have also been made by developing better bioinformatics tools for gRNA design and selection,20,29 varying gRNA lengths,30,31 and using inducible CRISPR-Cas98,32 as well as applying the CRISPR-Cas9 protein–gRNA complex directly.32 PAM alterations were also generated to broaden the genome coverage for different CRISPR-Cas9s.33-35 Pre-screening of gRNAs using different fluorescent reporters and T-7 endonuclease sensitivity assays in prior experiments have been successfully used to increase the specificity and efficacy of candidate target DNA sites.36-39 Nevertheless the focus of current studies, namely the development of a “super” variant of CRISPR-Cas9 with optimal specificity and efficiency for all genomic target DNA sites, is highly desirable but might not be achievable because of the great variations of genomic compositions and context among the target DNA sites. Therefore, a different strategy must be developed to address these issues. In this article, we will summarize the challenges and the most recent progresses in CRISPR-Cas9 technology as well as provide perspectives for potential future research directions to optimize genome editing through customizing CRISPR-Cas9 to the target protospacer DNA.
CRISPR-Cas9 is a finely tuned and safeguarded endonuclease
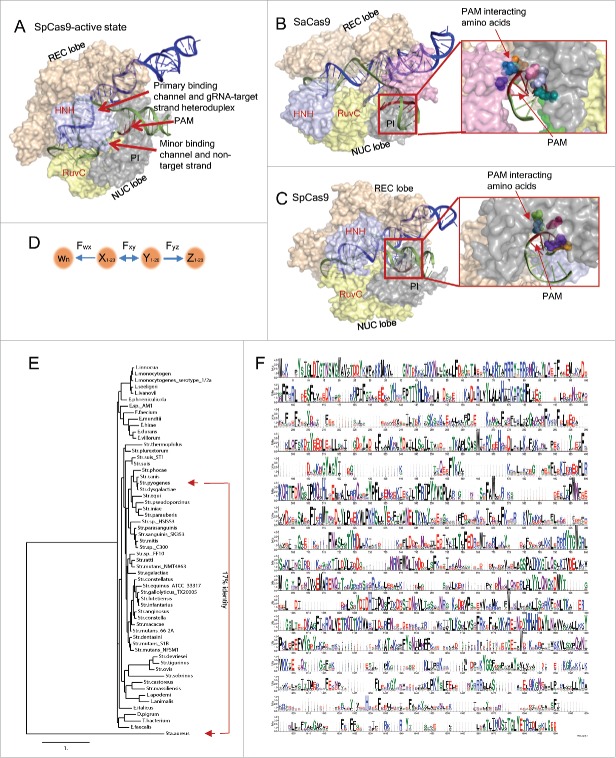
CRISPR-Cas9 uses a 20 nucleotide gRNA as a guide to find the complementary protospacer DNA target in a genome where it cuts the double stranded DNA precisely 3 base pairs upstream of the PAM sequence, a process that requires CRISPR-Cas9 to undergo several complicated but finely-tuned conformational changes (Fig. 1A–C).23-25,28,40 The CRISPR-Cas9 directed genome editing starts from the binding of a sgRNA (single guide RNA) to the primary binding channel formed between the REC and NUC lobes as well as other associated domains (Fig. 1A), which transforms the CRISPR-Cas9 from the inactive standby state to the active gRNA-directed target DNA search mode. Once the complementary target protospacer DNA is found, the CRISPR-Cas9-sgRNA complex binds to the protospacer DNA and with the help of the PI (PAM interacting) domain, the gRNA starts interrogating the double stranded DNA by base pairing with the target DNA strand from the PAM proximal position. The primary binding channel gradually adopts a new conformation to bind the gRNA-target DNA heteroduplex while the displaced non-target DNA strand is bound to the minor binding channel formed within the NUC lobe.23,40 As the base pairing process continues to the PAM distal end of the protospacer DNA, the HNH domain of CRISPR-Cas9 progressively makes about a 180° turn until it is perfectly positioned near the third nucleotide position of the target DNA strand proximal to the PAM. Meanwhile the RuvC-I domain is changed from the buried state to the exposed state and positioned at the same location of the non-target DNA strand just opposite to the HNH domain. Once both the HNH and RuvC-I domains are locked in the desired position, the nuclease activities of these 2 domains break both target and non-target strands of the DNA simultaneously, resulting in a so-called double strand DNA break (DSB). Thus, the CRISPR-Cas9 endonuclease activities are regulated at 3 levels: the first is the activation stage through the binding of sgRNA for target DNA searching and binding, which ensures CRISPR-Cas9 does not randomly bind DNA. The second is the repositioning of the HNH and RuvC-I nuclease domains to the expected position, which can only happen when the gRNA and target DNA heteroduplex is correctly formed to make sure only the selected DNA is targeted. The third is the DNA double strand cutting stage, which can only happen after the first and second stages are completed perfectly. This triple level secured activation mechanism of CRISPR-Cas9 nuclease activities guarantees that the sgRNA free CRISPR-Cas9 cannot bind DNA randomly and the DSB only occurs when the expected gRNA and target DNA heteroduplex is truly formed in order to prevent accidental cutting of unwanted off-target DNA.
Figure 1.
Schematic diagram of the functional mechanism and structural conservation of CRISPR-Cas9. (A) gRNA interrogation of target protospacer DNA created according to PDB ID:5F9R. The heteroduplex of gRNA and the target strand of the protospacer DNA is bound in the primary binding channel formed between the REC and NUC lobes; the non-target strand of the protospacer DNA is bound in the minor binding channel formed within the NUC lobe; the 2 HNH and RuvC endonuclease domains, the PI domain and PAM site are also indicated. (B) SaCas9 and gRNA complex highlighting the PAM and PAM interacting amino acids of the PI domain, created according to PDB ID: 5CZZ. Seven colors represent different amino acids. (C) SpCas9 and gRNA complex highlighting the PAM and PAM interacting amino acids of the PI domain created according to PDB ID: 4UN3. Five colors represent 5 different amino acids. (D) Interacting forces among the gRNA (Z), target DNA strand (Y), non-target DNA strand (X) and amino acids in the minor binding channel (Wn). Fyz represents hydrogen bonds of the pairing nucleotides between gRNA and target DNA strand of the protospacer DNA; Fxy represents the hydrogen bonds of the pairing nucleotides between the target and non-target strands of the protospacer DNA; Fwx represents the pulling force generated from the minor binding channel and asserting on the non-target strand of the protospacer DNA. (E) Phylogenetic tree of CRISPR-Cas9s (ConSurf and Phylogeny.fr) from 58 bacterial species showing distant evolutionary relations between SpCas9 and SaCas9. The 3 letters of the genus names were adopted only for the purpose of distinctions. (F) Weblogo display of the amino acid sequence homologies represents discrete homologous and non-homologous regions among the 58 CRISPR-Cas9s described in E. The color represents amino acid property and the size corresponds to relative degree of conservation.
The CRISPR-Cas9 promise
The greatest advantages of the CRISPR-Cas9 system are its simplicity and wide applicability in genome manipulations of almost all biological systems tested to date, including cell lines, stem cells, yeasts, worms, insects, rodents, and mammals. For a targetable DNA site, only a corresponding 20 nucleotide gRNA is needed to guide the CRISPR-Cas9 to cut the target DNA at the desired location. The repair of the broken DNA ends occurs either through NHEJ to generate indels, which has been used to generate random genomic mutations or through HDR in the presence of donor oligonucleotides or DNA fragments containing homologous sequences flanking the DSB sites to generate precise site-directed nucleotide or large gene replacements, leading to generation of targeted gene mutations or corrections.41,42 In addition to gene editing, dCas9 (dead Cas9) has been employed to manipulate whole genome gene expression profiles by either activating (CRISPRa) or inhibiting (CRISPRi) expression of 1 or more target genes simultaneously.8-10,43,44 The dCas9 has also been successfully used in whole genome screening of genes associated with cholera toxin sensitivity and epigenetic modifications.8 In order to study gene regulatory networks and genetic interactions scaffold RNAs (scRNAs) were developed to simultaneously activate or repress multiple genes in a single cell.45 More recently developed is a split dCas9-regulator fusion protein system whose activities are either blue light or chemically inducible in order to achieve spatial and temporal controls of dCas9 activities.8,11,13,18,46,47 These progresses illustrate the invaluable potentials of the CRISPR-Cas9 endonuclease in both gene editing and regulation at both cellular and organismal levels. However the most exciting application of the CRISPR-Cas9 technology is its potential in the treatment of human diseases. It has been shown that CRISPR-Cas9-mediated gene editing can be used for correction of genetic mutations in animals to cure genetic diseases and has been piloted in treatment of cancers, HIV and others.11-13,18,26,30,34,47 There have been a growing number of preclinical trials targeting various human diseases even though it is currently still at an imperfect stage for clinical applications.13 More recently, a high throughput platform (CombiGEM) for genome wide screening of genes and drugs against ovary cancer cell growth has been developed,48 which is likely to facilitate gene and drug discoveries in other types of cancers as well.
Challenges and current solutions of the CRISPR-Cas9 technology
Off-target activity
Although the CRISPR-Cas9 technology has been successfully applied in gene editing and regulation in a wide range of biological systems, it still faces 3 inherent challenges before it could possibly be used in medicine. The foremost challenge is the off-target activity. Because the CRISPR-Cas9 DSB activity is modulated solely by a single 20 nucleotide gRNA and is tolerant to some base pair mismatches between gRNA and the target DNA, there is a possibility that it could cut at genomic locations only partially complementary to the gRNA. These off-target activities vary among different target DNA sites because of the variations of their nucleotide compositions and genomic context. This has been a serious concern because, for clinical applications, any off-target activity could cause undesirable consequences and is not acceptable. To address this problem, a number of methods have been proposed, from selection of gRNAs through various bioinformatics tools to using shorter gRNA, and inducible CRISPR-Cas9 as well as CRISPR-Cas9 protein directly.12,26,30,34 These efforts have had limited success because the target DNA sequences and genetic context are different for each DNA target. The resolution of the crystal structures of several CRISPR-Cas9s revealed the molecular interaction mechanisms between gRNA-guided CRISPR-Cas9 and its DNA targets.17,23,24,26,28,49 The interrogation of gRNA with the double stranded target DNA starts through the assistance of the PI domain of CRISPR-Cas9 while the continuation of the unwinding of the double strand protospacer DNA and formation of the gRNA-target DNA heteroduplex requires the assistance of the NUC lobe to pull away the non-target DNA from the gRNA-target DNA heteroduplex. It has been demonstrated that the pulling force of the NUC lobe is due to the positive electric cloud formed along the minor binding channel. Alterations of the positively charged amino acids that change the positive electric density along the minor binding channel of NUC lobe also changes CRISPR-Cas9 DSB specificity. The strong pulling force due to the more positively charged amino acids along the minor binding channel weakens the hydrogen bonds between the 2 strands of the protospacer DNA, making it easier for the gRNA to displace the non-target DNA strand in order to form the gRNA-target DNA strand heteroduplex. With a balanced pulling force from the minor binding channel of the NUC lobe, it would require a more perfectly matched gRNA to displace the non-target DNA strand to form a heteroduplex with the target DNA strand. If the pulling force is too strong, it would more likely produce off-target DSB activities; likewise if the pulling force is too weak, even the perfectly matched gRNA could not displace the non-target DNA strand and the DSB efficiency would be compromised. Illustration of gRNA–target DNA interaction mechanisms opens the door for the rational design of a new version of CRISPR-Cas9 with improved specificities as demonstrated by SpCas9-HF1(Cas9 from Streptococcus pyogenes)21 and SpCas9 (1.0) and (1.1),26 each with different combinations of amino acid alterations. Thus, systematically identifying amino acids responsible for each base pair position of the protospacer sequence would allow targeted engineering of the CRISPR-Cas9 tailored to each target DNA site.
PAM restrictions
The precision of the CRISPR-Cas9 DSB activity requires the presence of PAM that serves as an essential site for CRISPR-Cas9-sgRNA complex landing at the protospacer DNA and initiating the target DNA interrogation. CRISPR-Cas9s from different bacterial species have been found to have different PAMs with various lengths and nucleotide compositions. Each type of PAM determines the cutting frequencies of the CRISPR-Cas9 for a given genome. None of the PAMs identified so far or even a combination of all of the known PAMs could possibly cover any whole genome sequences, which would in some cases restrict the use of CRISPR-Cas9 technology. To relax such PAM limitations, progresses have been made to modify the CRISPR-Cas9 PAMs by either bacterial selection-based directed evolution33,34 or through structure based rational design. Variants of the SpCas9 have been created to recognize different PAMs other than “NGG”, including 1 variant recognizing a 2 nucleotide “YG” PAM. The 2 nucleotide PAM would significantly increase genome editing coverage and broaden site-specific gene therapies.24 However too many PAMs nested on a target DNA is found inhibitive to in vivo CRISPR-Cas9 activities.50 The ideal strategy would be to customize a PAM sequence tailored only to the desired target DNA site through alteration of the responsible PAM interacting amino acids, which has been made possible by the identification of the amino acids responsible for PAM recognition in both SpCas9 and SaCas9 (Cas9 from Staphylococcus aureus) (Fig. 1B and C).
Variations of efficiency
The efficiency of CRISPR-Cas9 directed DSB activities varies widely depending on the nucleotide compositions and genomic context of the target protospacer DNA sites as well as the sgRNA secondary structure.17 Some gRNA sequence features have been identified to significantly affect the efficiency based on the analyses of more than 200 gRNAs. Although the general applicability of the discovery is waiting to be tested, it does highlight the significant role of gRNA nucleotide composition in efficiency. Similar results were also obtained in other independent studies, showing significant variations in efficiency between gRNAs targeting adjacent protospacer sites on the same plasmid. Unfortunately, until now, there was no reliable bioinformatics method to broadly predict the gRNA and CRISPR-Cas9 efficiencies at a target DNA site. Currently the efficiency of a gRNA at a specific DNA target can realistically only be experimentally determined. A number of technologies have been developed for screening gRNA efficiency and specificity, including both comprehensive technologies (NGS and GUIDE-Seq) and more simple methods (Surveyor assay and Fluorescent reporter assay21,26,37,38,39). Additionally the genome editing efficiency is also affected by NHEJ (higher) and HDR (lower) DNA repair mechanisms. HDR efficiencies can be increased by varying the insert/donor size, modifying DNA donors with phosphorothioation and inhibition of NHEJ activity.51 Recently, it was demonstrated that NHEJ can also be used to facilitate large DNA fragment (IRES-GFP) knock-in with higher efficiencies than HDR.52
Idealization of CRISPR-Cas9
Recent advances in different frontlines of CRISPR-Cas9 technology have opened up new avenues for further improvements in its specificity, efficiency, and genome coverage. The resolution of CRISPR-Cas9 crystal structures, especially using Cryo-EM technology to capture the CRISPR-Cas9/sgRNA/PAM- target DNA complex in action, provides details of its functional molecular mechanisms and the foundation for rational engineering of better CRISPR-Cas9.23,26,28 A comprehensive biophysical model was also developed for optimizing genome editing and gene regulations of CRISPR-Cas9 activities.43 However, most of the efforts for CRISPR-Cas9 improvement have been primarily focusing on a “one for all strategy”: to make a “super” version of CRISPR-Cas9 suitable for all target DNA, a desirable goal that might not be realistically achievable. According to current CRISPR-Cas9 function models, the amino acids along the minor binding channel formed within the NUC lobe are critical for its specificity and efficiency. Several amino acids have been identified that affect the specificity of CRISPR-Cas9 activity by directly interacting with the corresponding nucleotides at defined positions of the non-target protospacer DNA. It appears that for the nucleotides at each position of the protospacer DNA, there might be corresponding amino acids in the NUC minor binding channel with dominant interactions. This hypothesis is supported by the finding that variants of CRISPR-Cas9 with defined amino acid mutations showed different DSB efficiencies at both on- and off- target DNA sites.21,26 The gRNA that aims at the target DNA site whose nucleotide composition is balanced with the electric positivity along the minor binding channel of the NUC lobe would be most effective and specific. This explains why 2 gRNAs targeting adjacent target DNA sites showed different specificity and efficiency even though their genetic contexts are similar.39 Therefore it is apparent that the nucleotide composition of the target protospacer DNA plays an essential role in both efficiency and specificity. Approximately there are 4E+20 possible permutations for a 20 nucleotide protospacer DNA and only a small portion of them could possibly be balanced or close to being balanced with the electric positivity of the minor binding channels of a given CRISPR-Cas9 NUC lobe. Thus it seems unrealistic to create a CRISPR-Cas9 variant suitable for every possible composition of protospacer DNA.
For any given nucleotide position of the target protospacer DNA duplex, there are 3 interacting forces: the first force is the hydrogen-bonds between the opposing nucleotides of the DNA duplex (Fxy); the second force is from the nucleotide of the corresponding gRNA (Fyz) and the third force (Fwx) is from 1 or more positively charged amino acids in the NUC minor binding channel (Fig. 1D).21,26 The delicate balance of the 3 forces determines the fate of RNA-DNA heteroduplex formation. Because the gRNA and the target protospacer DNA interactions are predetermined by nucleotide composition, the only variable force is Fwx from the positively charged amino acids in the NUC minor binding channel. When Fwx is stronger, the non-target strand DNA would be pulled away harder from the DNA duplex, and the hydrogen bonds between the 2 protospacer DNA strands (Fxy) would become weaker, which would make it easier for the interrogating gRNA to displace the non-target strand DNA, consequently resulting in higher DSB efficiency. However if the Fwx is too strong, even the imperfectly matched gRNA could displace the non-target strand DNA, which would cause compromised specificity and off-target activities. On the other hand, if the Fwx is too weak, even perfectly matched gRNA could not break the hydrogen bonds of DNA-DNA duplex, and the DSB efficiency would be compromised. Therefore the fine tuning of the 3 interacting forces would be the key to achieving both high specificity and efficiency. To do this, the interacting forces for each nucleotide position of the target protospacer DNA must be defined by systematic mutagenesis or alanine scanning and corresponding crystal structure analysis. Once all 3 forces are known, the amino acid positivity could be adjusted according to the target protospacer DNA sequences. Ultimately, a customized CRISPR-Cas9 tailored to each nucleotide position of a target protospacer DNA site could be created through site directed mutagenesis.
PAM determines the genome locations where the CRISPR-Cas9/gRNA complex could land and initiate DNA interrogation by gRNA.23 The strict requirement of a special PAM sequence contributes to the DSB specificity but is also a primary factor restricting the genome editing frequency of a CRISPR-Cas9. Amino acids in the PI domains of both SpCas9 and SaCas9 have been identified to be responsible for PAM recognition. The longer the PAM sequences, the more corresponding amino acids of CRISPR-Cas9 are needed as revealed from both PAM binding sites of SpCas9 and SaCas9 (Fig. 1B and C).23,28 The SpCas9 that recognizes a 3 nucleotide PAM, NGG, requires 5 amino acids while SaCas9 that recognizes a 6 nucleotide PAM, NNGRRT, requires 7 interacting amino acids. More interestingly, alteration of the amino acids in the PI domain made the CRISPR-Cas9 recognize different PAM sequences with equal or better DSB activities.33-35 Thus, it is also possible to engineer the CRISPR-Cas9 to recognize a novel PAM sequence at a desired target DNA location. With the knowledge of the CRISPR-Cas9 crystal structure in action and standardized site-directed mutagenesis protocols, it is feasible to custom design a CRISPR-Cas9 that can recognize a unique PAM sequence tailored to a specific DNA target and every target in a genome by alteration of the corresponding amino acids of the CRISPR-Cas9 PI domain to maximize the specificity and genome editing coverage.
In addition to customizing the NUC minor binding channel and PAM interacting amino acids, the CRISPR-Cas9 system could be further improved by shrinking its size. Current size of Cas9 sequences has limitations for clinically promising vectors, such as adeno-associated virus (AAV), which can only accomodate an insert size up to 4.5kb. The SpCas9 is about 4.2 kb in size, making it difficult to create the integrated Cas9-sgRNA AAV constructs. The SaCas9 that showed similar functional features as SpCas9, but is only about 3 kb in size, would allow more room for integration of both CRISPR-Cas9 and 1 or more sgRNAs into a single AAV vector. The crystal structures of both CRISPR-Cas9 proteins showed similar domain and structure configurations (Fig. 1B and C), indicating there might be more non-essential sequences existing in the CRISPR-Cas9 sequences. Functional homology searches of SpCas9 using ConSurf and phylogenetic analysis, identified variants of CRISPR-Cas9 from 58 bacterial species that have similar function domains, but various homologies.53,54 SaCas9 is one of the most distant variants from SpCas9, sharing only 17% overall identity (Fig. 1E). Although both CRISPR-Cas9s and others in alignment share discrete highly conserved stretches of sequences including RuvC-1 domain and the HNH domains, there are also stretches of regions that are low in homology and might be disposable (Fig. 1F).55 These variations strongly suggest that the CRISPR-Cas9s from different resources are structurally and functionally conserved, but vary at the level of amino acid sequences, which make it possible to design an artificial CRISPR-Cas9 with better function and smaller size.
To summarize, the medical potential of CRISPR-Cas9 technology could best be realized by employing customized variants to specifically suit the target DNA site. The ideal application process of genome editing in the future is proposed as below: 1) identify the desirable target DNA site; 2) determine the PAM sequence so that the protospacer sequence is unique to the genome of interest; 3) calculate the amino acid positivity of the NUC minor binding channel needed for each position of the protospacer sequence; 4) determine what amino acids in the minor binding channel of the NUC lobe need to be replaced with desirable amino acids for each nucleotide position of the target protospacer DNA; 5) engineer the template CRISPR-Cas9 to the desired amino acid sequence by site-directed mutagenesis or gene synthesis; 6) test the specificity and efficiency of different variants of CRISPR-Cas9 in vitro using different fluorescent reporters to select those with best efficiency and specificity; 7) test the activities of the candidate variants of CRISPR-Cas9 in vivo using desired animal models; 8) pre-clinical and clinical trials of the winning customized CRISPR-Cas9-gRNA complexes.
There is still a long way to go in order to reach the proposed status for CRISPR-Cas9 technology. But with the unparalleled enthusiasm from scientists in both structural and experimental biology as well as other related fields, it is entirely plausible to predict that the perfected version of the CRISPR-Cas9 is within reach.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
References
- [1].Weninger A, Hatzl AM, Schmid C, Vogl T, Glieder A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J Biotechnol. 2016 Mar 22. PII: S0168-1656(16)30134-1. http://dx.doi.org/ 10.1016/j.jbiotec.2016.03.027. [DOI] [PubMed] [Google Scholar]
- [2].Calarco JA, Friedland AE. Creating genome modifications in C. elegans Using the CRISPR/Cas9 System. Methods Mol Biol 2015;1327:59-74; PMID:26423968; http://dx.doi.org/ 10.1007/978-1-4939-2842-2_6 [DOI] [PubMed] [Google Scholar]
- [3].Housden BE, Lin S, Perrimon N. Cas9-based genome editing in Drosophila. Methods Enzymol 2014;546:415-39; PMID:25398351; http://dx.doi.org/ 10.1016/B978-0-12-801185-0.00019-2 [DOI] [PubMed] [Google Scholar]
- [4].Hu X, Wang C, Fu Y, Liu Q, Jiao X, Wang K. Expanding the range of CRISPR/Cas9 genome editing in rice. Mol Plant 2016. [DOI] [PubMed] [Google Scholar]
- [5].Maresch R, Mueller S, Veltkamp C, Ollinger R, Friedrich M, Heid I, Steiger K, Weber J, Engleitner T, Barenboim M, et al.. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun 2016;7:10770; PMID:26916719; http://dx.doi.org/ 10.1038/ncomms10770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kang Y, Zheng B, Shen B, Chen Y, Wang L, Wang J, Niu Y, Cui Y, Zhou J, Wang H, et al.. CRISPR/Cas9-mediated Dax1 knockout in the monkey recapitulates human AHC-HH. Hum Mol Genet 2015;24:7255-64; PMID:26464492; http://dx.doi.org/ 10.1093/hmg/ddv425 [DOI] [PubMed] [Google Scholar]
- [7].Graham DM. CRISPR/Cas9 corrects retinal dystrophy in rats. Lab Animal 2016;45:85; PMID:26886660; http://dx.doi.org/ 10.1038/laban.965 [DOI] [PubMed] [Google Scholar]
- [8].Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 2016;17:5-15; PMID:26670017; http://dx.doi.org/ 10.1038/nrm.2015.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, Klasic M, Zoldos V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 2016; PMID:26969735; http://dx.doi.org/ 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pham H, Kearns NA, Maehr R. Transcriptional regulation with CRISPR/Cas9 effectors in mammalian cells. Methods Mol Biol 2016;1358:43-57; PMID:26463376; http://dx.doi.org/ 10.1007/978-1-4939-3067-8_3 [DOI] [PubMed] [Google Scholar]
- [11].Zhang N, Zhi H, Curtis BR, Rao S, Jobaliya C, Poncz M, French DL, Newman PJ. CRISPR/Cas9-mediated conversion of human platelet alloantigen allotypes. Blood 2016;127:675-80; PMID:26634302; http://dx.doi.org/ 10.1182/blood-2015-10-675751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol Ther: J Am Soc Gene Ther 2016;24:636-44; PMID:26658966; http://dx.doi.org/ 10.1038/mt.2015.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Savic N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Translat Res: J Lab Clin Med 2016;168:15-21; PMID:26470680; http://dx.doi.org/ 10.1016/j.trsl.2015.09.008 [DOI] [PubMed] [Google Scholar]
- [14].Shinmyo Y, Tanaka S, Tsunoda S, Hosomichi K, Tajima A, Kawasaki H. CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci Rep 2016;6:20611; PMID:26857612; http://dx.doi.org/ 10.1038/srep20611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vassena R, Heindryckx B, Peco R, Pennings G, Raya A, Sermon K, Veiga A. Genome engineering through CRISPR/Cas9 technology in the human germline and pluripotent stem cells. Hum Reproduct Update 2016; PMID:26932460; http://dx.doi.org/ 10.1093/humupd/dmw005 [DOI] [PubMed] [Google Scholar]
- [16].Wang D, Ma N, Hui Y, Gao X. [The application of CRISPR/Cas9 genome editing technology in cancer research]. Yi chuan = Hereditas / Zhongguo yi chuan xue hui bian ji 2016;38:1-8. [DOI] [PubMed] [Google Scholar]
- [17].Liu X, Homma A, Sayadi J, Yang S, Ohashi J, Takumi T. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci Rep 2016;6:19675; PMID:26813419; http://dx.doi.org/ 10.1038/srep19675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].White MK, Khalili K. CRISPR/Cas9 and cancer targets: future possibilities and present challenges. Oncotarget 2016;7:12305-17; PMID:26840090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Renaud JB, Boix C, Charpentier M, De Cian A, Cochennec J, Duvernois-Berthet E, Perrouault L, Tesson L, Edouard J, Thinard R, et al.. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep 2016;14:2263-72; PMID:26923600; http://dx.doi.org/ 10.1016/j.celrep.2016.02.018 [DOI] [PubMed] [Google Scholar]
- [20].Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al.. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184-91; PMID:26780180; http://dx.doi.org/ 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016;529:490-5; PMID:26735016; http://dx.doi.org/ 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen H, Bailey S. Structural biology. Cas9, poised for DNA cleavage. Science 2016;351:811-2; PMID:26912877; http://dx.doi.org/ 10.1126/science.aaf2089 [DOI] [PubMed] [Google Scholar]
- [23].Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016;351:867-71; PMID:26841432; http://dx.doi.org/ 10.1126/science.aad8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, et al.. Structure and Engineering of Francisella novicida Cas9. Cell 2016;164:950-61; PMID:26875867; http://dx.doi.org/ 10.1016/j.cell.2016.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zlotorynski E. Genome engineering: Structure-guided improvement of Cas9 specificity. Nat Rev Mol Cell Biol 2016;17:3. [Google Scholar]
- [26].Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016;351:84-8; PMID:26628643; http://dx.doi.org/ 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nelson CE, Gersbach CA. Cas9 loosens its grip on off-target sites. Nat Biotechnol 2016;34:298-9; PMID:26963555; http://dx.doi.org/ 10.1038/nbt.3501 [DOI] [PubMed] [Google Scholar]
- [28].Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell 2015;162:1113-26; PMID:26317473; http://dx.doi.org/ 10.1016/j.cell.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brazelton VA Jr., Zarecor S, Wright DA, Wang Y, Liu J, Chen K, Yang B, Lawrence-Dill CJ. A quick guide to CRISPR sgRNA design tools. GM Crops Food 2015;6:266-76; PMID:26745836; http://dx.doi.org/ 10.1080/21645698.2015.1137690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 2013;31:839-43; PMID:23934178; http://dx.doi.org/ 10.1038/nbt.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al.. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015;520:186-91; PMID:25830891; http://dx.doi.org/ 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PloS One 2014;9:e98186; PMID:24873830; http://dx.doi.org/ 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 2015;33:1293-8; PMID:26524662; http://dx.doi.org/ 10.1038/nbt.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al.. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015;523:481-5; PMID:26098369; http://dx.doi.org/ 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li Y, Mendiratta S, Ehrhardt K, Kashyap N, White MA, Bleris L. Exploiting the CRISPR/Cas9 PAM constraint for single-nucleotide resolution interventions. PloS One 2016;11:e0144970; PMID:26788852; http://dx.doi.org/ 10.1371/journal.pone.0144970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jinqing W, Gui M, Zhiguo L, Yaosheng C, Peiqing C, Zuyong H. Improving gene targeting efficiency on pig IGF2 mediated by ZFNs and CRISPR/Cas9 by using SSA reporter system. Yi chuan = Hereditas / Zhongguo yi chuan xue hui bian ji 2015;37:55-62. [DOI] [PubMed] [Google Scholar]
- [37].Ramakrishna S, Cho SW, Kim S, Song M, Gopalappa R, Kim JS, Kim H. Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat Commun 2014;5:3378; PMID:24569644; http://dx.doi.org/ 10.1038/ncomms4378 [DOI] [PubMed] [Google Scholar]
- [38].Vriend LE, Jasin M, Krawczyk PM. Assaying break and nick-induced homologous recombination in mammalian cells using the DR-GFP reporter and Cas9 nucleases. Methods Enzymol 2014;546:175-91; PMID:25398341; http://dx.doi.org/ 10.1016/B978-0-12-801185-0.00009-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang JH, Pandey M, Kahler JF, Loshakov A, Harris B, Dagur PK, Mo YY, Simonds WF. Improving the specificity and efficacy of CRISPR/CAS9 and gRNA through target specific DNA reporter. J Biotechnol 2014;189:1-8; PMID:25193712; http://dx.doi.org/ 10.1016/j.jbiotec.2014.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. Structural biology. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 2015;348:1477-81; PMID:26113724; http://dx.doi.org/ 10.1126/science.aab1452 [DOI] [PubMed] [Google Scholar]
- [41].Quetier F. The CRISPR-Cas9 technology: Closer to the ultimate toolkit for targeted genome editing. Plant Sci: Int J Exp Plant Biol 2016;242:65-76; PMID:26566825; http://dx.doi.org/ 10.1016/j.plantsci.2015.09.003 [DOI] [PubMed] [Google Scholar]
- [42].Urnov F. Genome editing: The domestication of Cas9. Nature 2016;529:468-9; PMID:26819037; http://dx.doi.org/ 10.1038/529468a [DOI] [PubMed] [Google Scholar]
- [43].Farasat I, Salis HM. A biophysical model of CRISPR/Cas9 activity for rational design of genome editing and gene regulation. PLoS Comput Biol 2016;12:e1004724; PMID:26824432; http://dx.doi.org/ 10.1371/journal.pcbi.1004724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xue HY, Ji LJ, Gao AM, Liu P, He JD, Lu XJ. CRISPR-Cas9 for medical genetic screens: applications and future perspectives. J Medical Genet 2016;53:91-7; PMID:26673779 [DOI] [PubMed] [Google Scholar]
- [45].Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al.. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015;160:339-50; PMID:25533786; http://dx.doi.org/ 10.1016/j.cell.2014.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 2015;11:198-200; PMID:25664691; http://dx.doi.org/ 10.1038/nchembio.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wojtal D, Kemaladewi DU, Malam Z, Abdullah S, Wong TW, Hyatt E, Baghestani Z, Pereira S, Stavropoulos J, Mouly V, et al.. Spell checking nature: versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am J Hum Genet 2016;98:90-101; PMID:26686765; http://dx.doi.org/ 10.1016/j.ajhg.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wong AS, Choi GC, Cui CH, Pregernig G, Milani P, Adam M, Perli SD, Kazer SW, Gaillard A, Hermann M, et al.. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci U S A 2016;113:2544-9; PMID:26864203; http://dx.doi.org/ 10.1073/pnas.1517883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shao S, Zhang W, Hu H, Xue B, Qin J, Sun C, Sun Y, Wei W, Sun Y. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucl. Acids Res. (19 May 2016) 44 (9): e86. http://dx.doi.org/ 10.1093/nar/gkw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Malina A, Cameron CJ, Robert F, Blanchette M, Dostie J, Pelletier J. PAM multiplicity marks genomic target sites as inhibitory to CRISPR-Cas9 editing. Nat Commun 2015;6:10124; PMID:26644285; http://dx.doi.org/ 10.1038/ncomms10124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 2016;7:10431; PMID:26786405; http://dx.doi.org/ 10.1038/ncomms10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].He X, Tan C, Wang F, Wang Y, Zhou R, Cui D, You W, Zhao H, Ren J, Feng B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016 May 19;44(9):e85. http://dx.doi.org/ 10.1093/nar/gkw064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 2010;38:W529-33; PMID:20478830; http://dx.doi.org/ 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al.. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008;36:W465-9; PMID:18424797; http://dx.doi.org/ 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res 2004;14:1188-90; PMID:15173120; http://dx.doi.org/ 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]