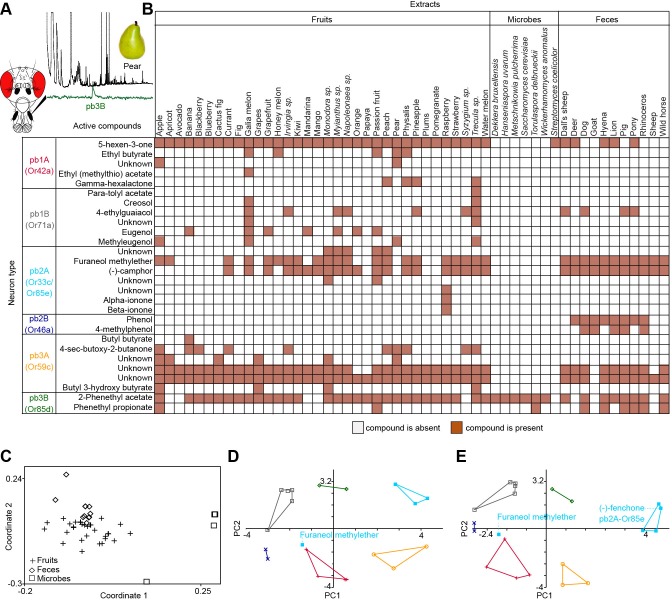
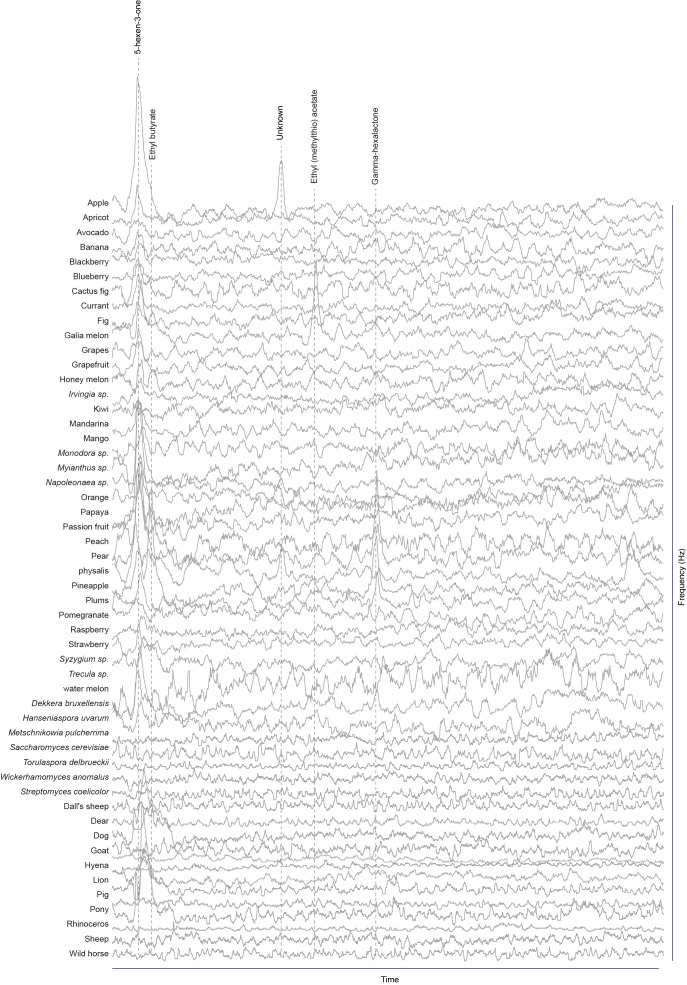
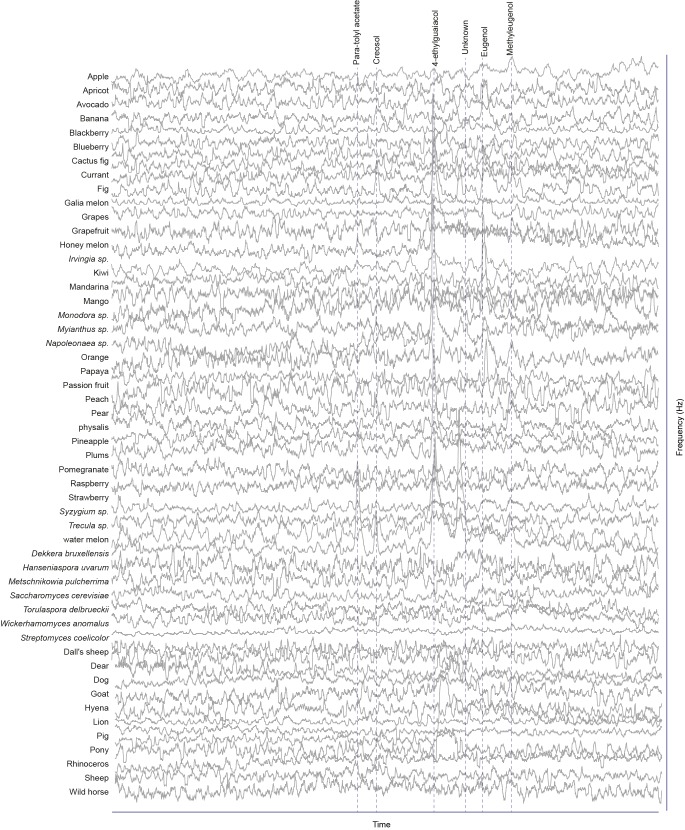
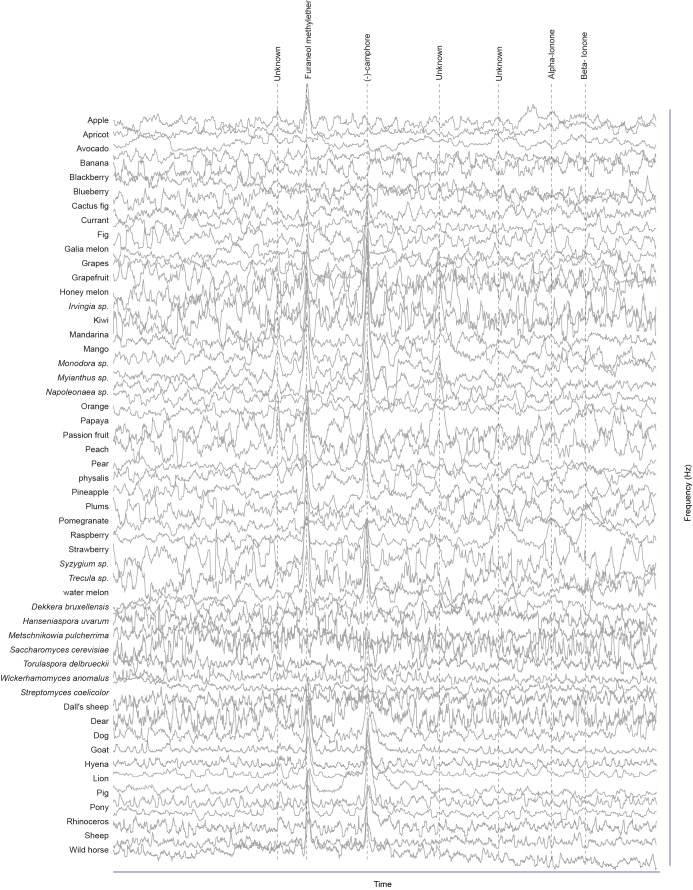
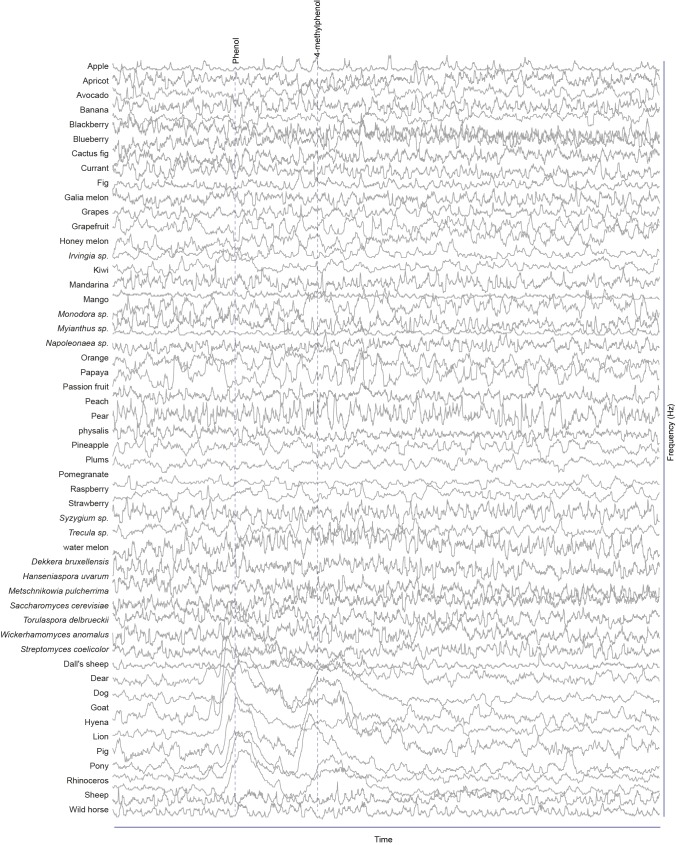
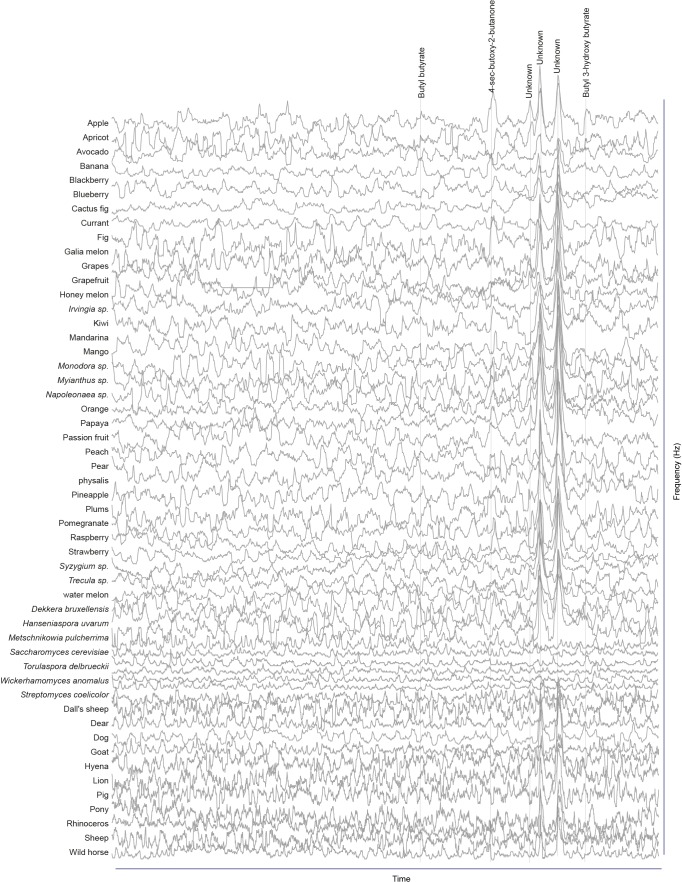
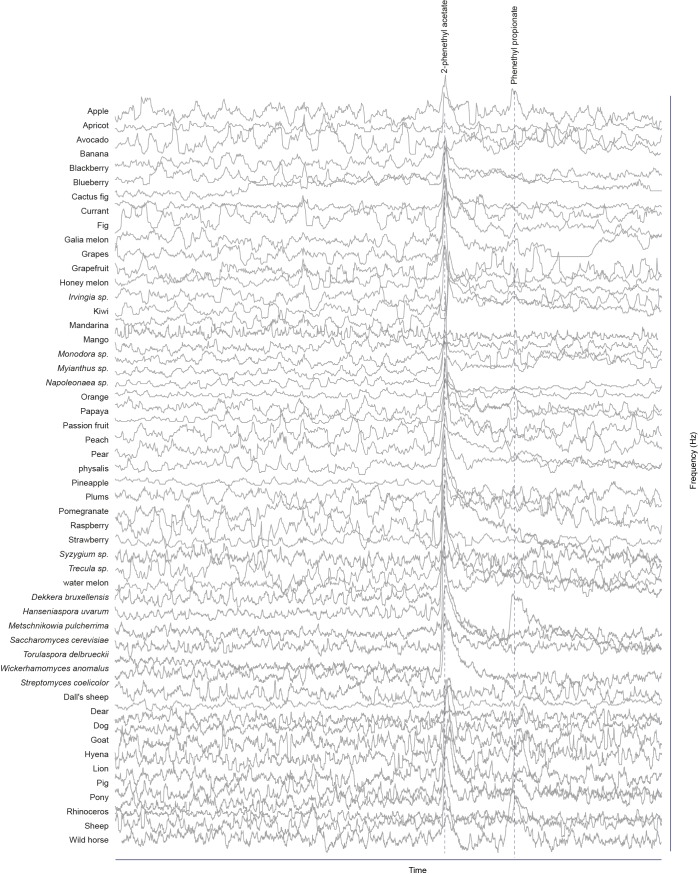
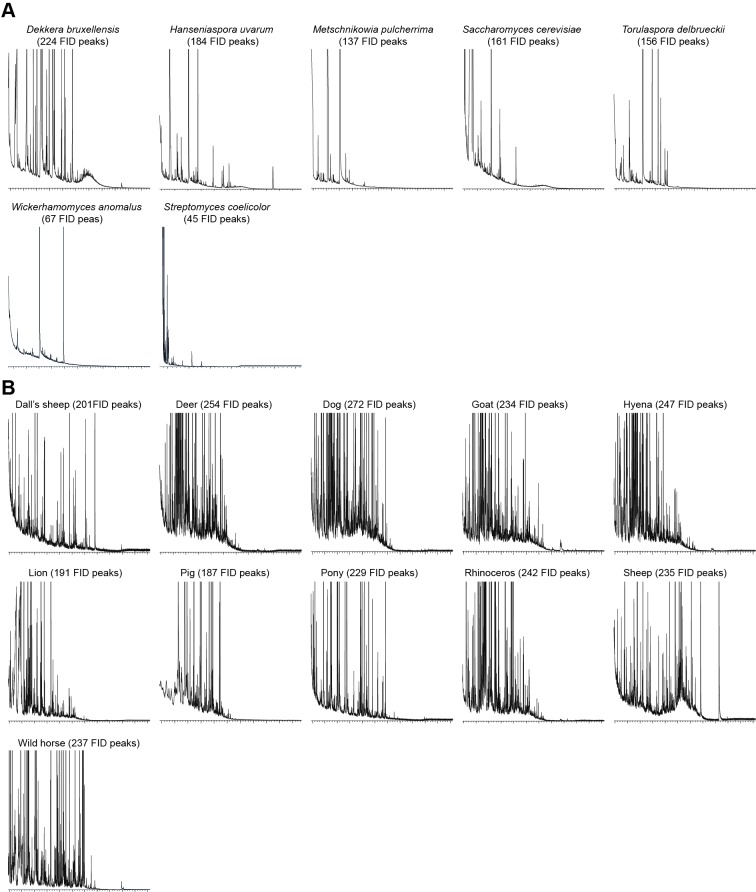
Figure 1. Screen for novel natural ligands for MP-OSNs.
(A) Representative gas chromatography-linked single sensillum measurement (GC-SSR) from pb3B (green trace) stimulated with headspace extract of pear (black trace). (B) Presence/Absence matrix of the physiologically active compounds identified from the different headspace extracts for each MP-OSN in the GC-SSR experiments (i.e. each filled box represents not only the presence of this odor in a specific fruit, but also a physiological response in GC-SSR recordings). (C) NMDS plot based on a presence/absence matrix for the active peaks across the tested samples. (D) PCA plot showing the distribution of the ligands recognized by MP-OSNs in a 32-dimensional odor space. PC1 and PC2 explain 23% and 22% of the variance, respectively. (E) PCA plot showing the distribution of the ligands recognized by MP-OSNs and (-)-fenchone (the main ligand of Or85e-expressing OSNs) in a 32-dimensional odor space. PC1 and PC2 explain 24% and 21% of the variance, respectively.
DOI: http://dx.doi.org/10.7554/eLife.14925.002