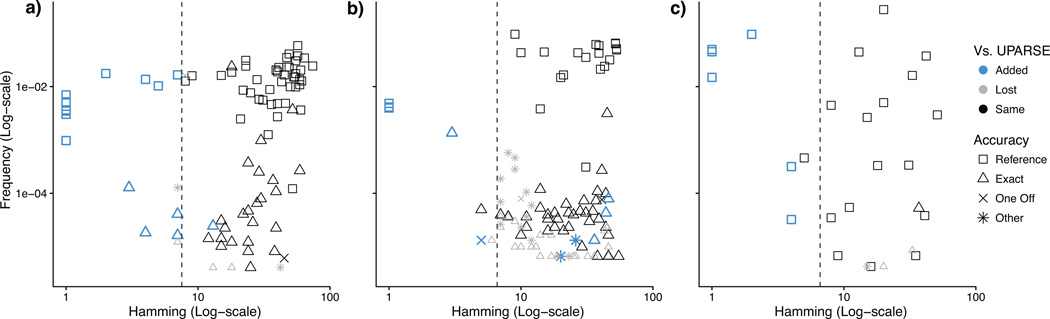
Figure 1. Sequence variants inferred by DADA2 compared to the OTUs constructed by UPARSE.
The merged sequences output by DADA2 are plotted for three Illumina amplicon datasets: (a) Balanced, (b) HMP, and (c) Extreme. Frequency is plotted on the y-axis; Hamming distance to the closest more-abundant sequence on the x-axis. Shapes represent accuracy (Methods). When variants are well separated from other members of the community the sequence variants inferred by DADA2 largely coincide with the OTUs output by UPARSE (black). However, DADA2 resolves additional variation (blue), especially within the UPARSE's OTU radius (dashed line), while outputting fewer spurious sequences (One Off and Other).