Fig. 1.
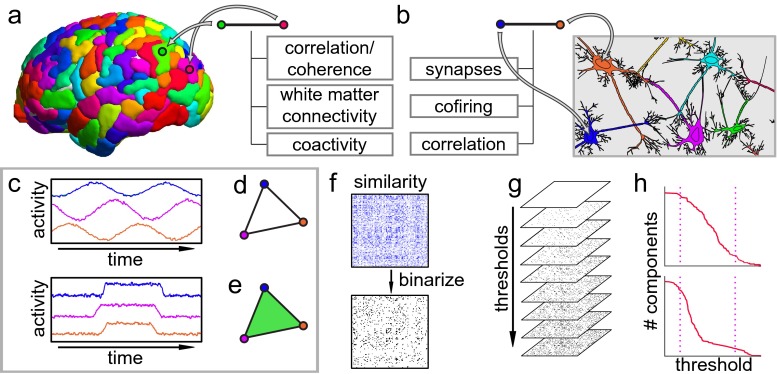
Extensions of network models provide insights into neural data. a Network models are increasingly common for the study of whole-brain activity. b Neuron-level networks have been a driving force in the adoption of network techniques in neuroscience. c Two potential activity traces for a trio of neural units. (top) Activity for a “pacemaker”-like circuit, whose elements are pairwise active in all combinations but never as a triple. (bottom) Activity for units driven by a common strong stimulus, thus are simultaneously coactive. d A network representation of the coactivity patterns for either population in (c). Networks are capable of encoding only dyadic relationships, so do not capture the difference between these two populations. e A simplicial complex model is capable of encoding higher order interactions, thus distinguishing between the top and bottom panels in (c). f A similarity measure for elements in a large neural population is encoded as a matrix, thought of as the adjacency matrix for a complete, weighted network, and binarized using some threshold to simplify quantitative analysis of the system. In the absence of complete understanding of a system, it is difficult or impossible to make a principled choice of threshold value. g A filtration of networks is obtained by thresholding at every possible entry and arranging the resulting family of networks along an axis at their threshold values. This structure discards no information from the original weighted network. g Graphs of the number of connected components as a function of threshold value for two networks reveals differences in their structure: (top) homogeneous network versus (bottom) a modular network. (dotted lines) Thresholding near these values would suggest inaccurately that these two networks have similar structure