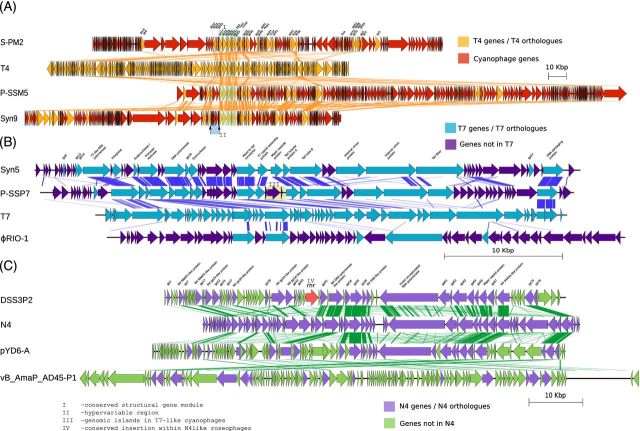
Figure 3.
Genomic comparison of selected phages. (A) Cyanophage S-PM2, P-SSM5 and Syn9 compared to phage T4. Orange genes are those found in phage T4. Blue shading is a hypervariable region localised between gp16 and gp17 in Syn9 (Millard et al.2009). The structural gene module that is common to all T4likeviruses is highlighted in green (Hambly et al.2001). (B) T7likevirus comparison of cyanophages Syn5 and P-SSP7, Pseudoalteromonas phage RIO-1 with phage T7. T7 genes found in other phages are teal coloured. The genomic islands identified in P-SSP7 are highlighted in yellow (Labrie et al.2013). (C) N4likevirus comparison of roseophage DSS3P2, Pseudoalteromonas phage pYD6-A and Alteromonas phage vB_AmaP_AD45-P1 with phage N4. Roseophage N4-like phages are represented by DSS3P2, which has genes that have been acquired by roseophages and localised at the same position (highlighted in red) (Chan et al.2014).