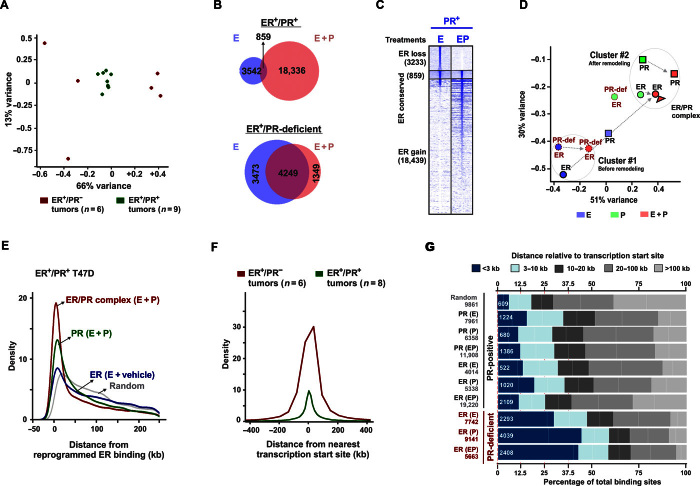
Fig. 2. PR redirects ER binding to sites correlated with the binding of PR and ER/PR complexes.
(A) Principal component analysis (PCA) plot displays 79% total variance between ER binding events in nine ER+/PR+ (green) and six ER+/PR− (red) patient tumors [sequencing data were obtained from the study by Ross-Innes et al. (27)]. (B) Overlap of ER binding sites in PR+ and PR-deficient T47D cells treated with estrogen or EP. (C) Heat maps display intensity of sequencing obtained on anti-ER ChIP before and after remodeling by R5020 in PR+ T47D cells. The genomic window of the union of all ER binding sites observed before and after remodeling by PR is displayed. Overlap of at least 1 base pair (bp) was considered to categorize ER binding as lost, conserved, or gained. (D) PCA plot depicts 81% total variance between binding events for ER, PR, and ER/PR complexes observed upon treatment with estrogen, progestin, or EP. Binding events in PR+ and PR-deficient T47D cells are presented. All the binding sites and their annotations are provided in table S6. (E) Distributions of receptor binding around reprogrammed ER binding sites. Distributions for ER binding observed without progestin and binding for PR and ER/PR complexes on estrogen plus progestin treatment are plotted. (F) Distributions around transcription start sites for ER binding observed in six ER+PR− and eight ER+PR+ human tumors. One outlier within the ER+/PR+ group was not included in the analyses. (G) Frequencies of binding events for ER, PR, and ER/PR complexes relative to their distance from transcription start sites. Hormone treatment is mentioned in parentheses. The numerical values for the total number of binding sites (gray) and the number of binding sites within the 3-kb promoter regions (white) are provided. ChIP-PCR assessments of ChIP-seq were done as three independent experiments and three technical replicates per experiment (fig. S4). ChIP-seq was performed on one of the three biological replicates.