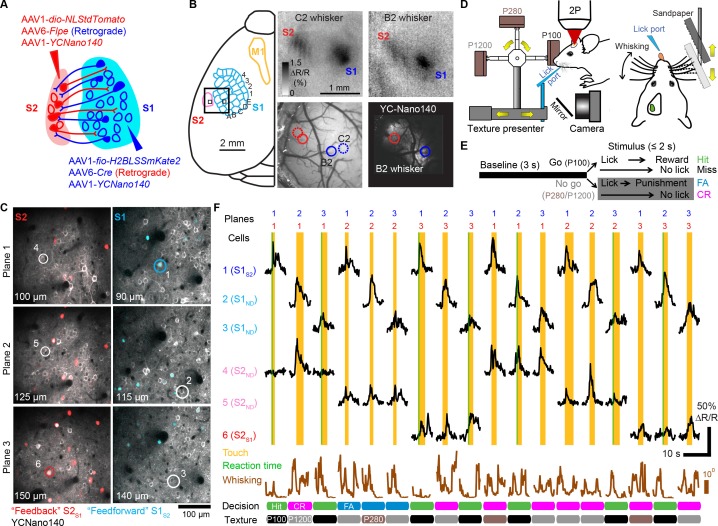
Figure 2. Simultaneous calcium imaging of identified feedforward and feedback neurons in S1 and S2 of mouse neocortex during behavior.
(A) Viral injection scheme for simultaneous labeling of feedforward and feedback neurons and YC-Nano140 expression. (B) Functional mapping of S1 and S2 through optical intrinsic signal imaging. Intrinsic signals evoked by stimulation of the C2 whisker (top left) and the B2 whisker (top right). In addition to localized intrinsic signals in S1 barrel columns, additional activation spots are visible in S2. Identified barrel columns (circles) are overlaid over blood vessel (bottom left) and YC-Nano140 expression (bottom right) images. (C) In vivo 2-photon images of LSSmKate2-positive S1S2 neurons (blue), tdTomato-positive S2S1 neurons (red) with non-co-labeled YC-Nano140-expressing neurons (grey) in S1 (S1ND) and S2 (S2ND). (D) Behavior setup for texture discrimination task. (E) Trial structure for go/no-go texture discrimination task. (F) Example calcium transients for individual neurons in [C] measured episodically during texture discrimination task along with periods of whisker-to-texture touch (orange area), whisking amplitude (brown trace), and reaction time on Hit trials (green area). For each trial the selected plane in each sub-area is indicated on top, illustrating the combinatorial plane hopping.
DOI: http://dx.doi.org/10.7554/eLife.14679.007