|
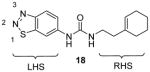
Assay: SAHH-coupled assay; SPA; RFA
IC50 1.6 μM (with H4-24 peptide as substrate), 2 μM (with rpS2 protein as substrate); Kd = 9 μM; inactive to five HKMTs (G9a, EHMT1, SUV39H2, SETD7, and SETD8), three PRMTs (-1, -4, and -8), weakly inhibit PRMT5; Allosteric binding, crystal structure available; Not metabolically stable enough for cell-based assays;
Note: all the methylation assays were done under balanced condition. |
2012[96] |
|
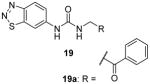
Assay: SPA; RFA
19a:
IC50 0.48 μM on PRMT3; 40-fold selective for PRMT3 over G9a, GLP, and SUV39H2; totally inactive against PRMT5, SETD7, PRC2, SETD8, SETDB1, SUV420H1, SUV420H2, MLL1, SMYD3, SMYD2, DOT1L, and DNMT1; Crystal structure available.
|
2013[80] |
|
Biochemical |
Cellular & Animal model |
|
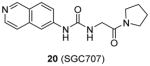
Assay: SPA
IC50 0.03 μM (PRMT3); outstanding selectivity against 31 HKMTs, PRMTs, DNMTs and RNA-methyltransferase as well as 250 non-epigenetic proteins; Crystal structure available; Kd = 0.053 μM and 0.085 μM, determined by ITC and SPR assay, respectively.
|
Bind with PRMT3 overexpressed in HEK293 and A549 cells (EC50 = 1.3 μM and 1.6 μM, respectively) determined by InCELL Hunter assay; Inhibit endogenous and exogenous H4R3me2a with (IC50 = 0.225μM and 0.091μM, respectively) from western blot; No toxicity at 24h; In vivo PK: The plasma level at 6 h post injection was 208 nm, more than 2-fold higher than its IC50 value in the cellular assay and the half-life was about 1 h.
|
2015[97] |



