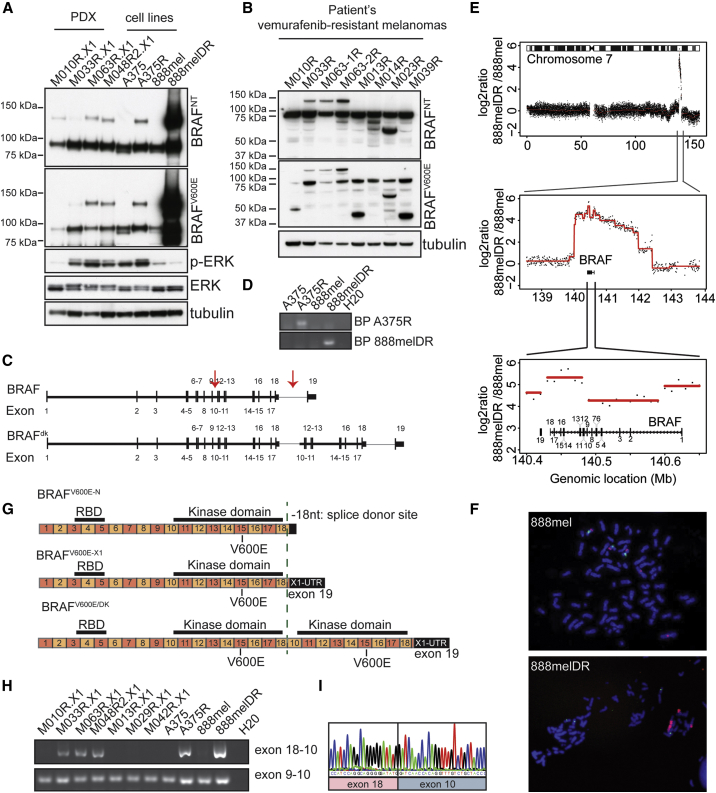
Figure 6.
Resistance Mechanism to BRAFi Involving Duplication of the Kinase Domain of BRAFV600E Discovered in PDX Panel
(A) Immunoblotting for BRAF using a set of four PDXs and two in vitro generated melanoma cell lines resistant for PLX4720 (A375R) or dabrafenib/trametinib (888melDR). Tubulin is used as a loading control. BRAFNT, antibody recognizing an epitope encoded by exons 2–3 of BRAF; BRAFV600E, antibody recognizing specifically the BRAFV600E epitope.
(B) Immunoblotting for BRAF on patient samples of vemurafenib-resistant lesions is shown.
(C) Representation of BRAFV600E and BRAFV600E/DK at the genomic level. Arrows indicate the introns where the breakpoints are localized. Black vertical bars, exons; horizontal bars, introns.
(D) Validation of specific genomic breakpoints is shown.
(E) DNA copy-number alterations in the 888melDR relative to 888mel cell line for chromosome 7, with magnification of the amplified region and further magnification of the BRAF locus, are shown.
(F) Fluorescence in situ hybridization (FISH) of 888mel and 888melDR cell line, using either a BRAF probe (red) or a chromosome 7 centromere probe (green), is shown.
(G) Illustration of BRAFV600E- and BRAFV600E/DK-encoding mRNA. Upper row indicates BRAFV600E with normal 3′ UTR (BRAFV600E−N), middle row indicates BRAFV600E with alternative X1 3′ UTR (BRAFV600E−X1), and bottom row indicates BRAFV600E/DK. Green dashed line indicates splice donor site localized within exon 18, which can be used for alternative 3′ UTR splicing.
(H) PCR product using a forward primer in exon 18 and a reverse primer in exon 10 validates the presence of BRAFV600E/DK. As a control, primers in exon 9 (forward) and exon 10 (reverse) were used.
(I) Sanger sequencing of PCR product obtained in (H) is shown.