Figure 1.
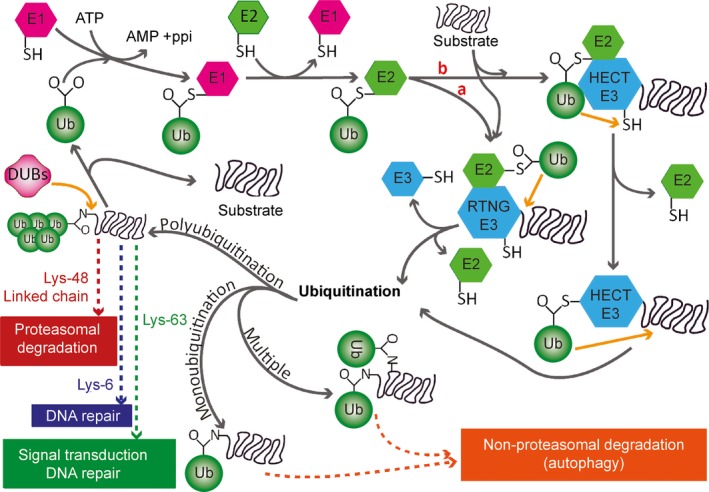
General outline of ubiquitination. Ubiquitination is the covalent attachment of the glycine residue of ubiquitin to the lysine residue of the target protein via an isopeptide bond. This process is mediated by E1, E2 and E3 enzymes. E1 ligases forms thioester bonds with ubiquitin, which is subsequently transferred to E2 ligases through a thioester linkage. E3 ligases are grouped into two types: HECT type and RING‐finger/adaptor type. Only the HECT domain E3 ligase forms a covalent bond with ubiquitin during polyubiquitination of abnormal proteins, whereas the RING‐finger E3 ligases directly transfer ubiquitin from the associated E2 enzyme to the substrate protein. Proteins can be mono‐, multiple‐ or poly‐ubiquitinated. Mono‐ and multiple‐ubiquitinated proteins are degraded by lysosomes (autophagy), whereas the degradation of polyubiquitinated proteins depends on the type of ubiquitin linkage (K48, K63, K6, K11, K27, K29 and K33). K48‐linked polyubiquitinated proteins are generally degraded by the proteasomal pathway. Polyubiquitination via K6 is associated with DNA repair; via K11, with endoplasmic reticulum‐associated protein degradation and cell cycle regulation; via K27, with ubiquitin fusion degradation; via K29, with lysosomal degradation; and via K33, with kinase modification. DUBs remove ubiquitin from the substrate protein, and the monomers are recycled.
