Figure 4.
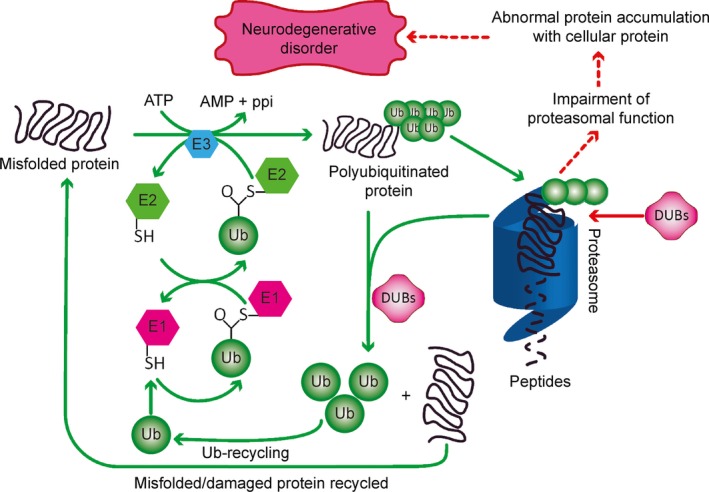
Delineation of the ubiquitin proteasome pathway in the pathogenesis of Alzheimer's disease. Improperly folded proteins are initially polyubiquitinated by the ubiquitin (Ub) enzymes E1 (Ub activating), E2 (Ub ligating) and E3 (Ub conjugating). These polyubiquitinated substrates are easily recognized by the 19S regulatory particle of the 26S proteasome. The polyubiquitinated protein is deubiquitinated by deubiquitinase enzymes (DUBs) and degraded into small peptides through 20S core particle. The polyubiquitinated proteins are deubiquitinated before they are recognized by the 26S proteasome. After they are recognized, they are polyubiquitinated again by the same mechanism and degraded by the 26S proteasome. DUBs convert poly‐Ub chains to Ub monomers, which are then recycled. Proteasome impairment decreases the degradation of amyloid beta (Aβ) and tau and promotes their oligomerization. The oligomers accumulate in neurons, resulting in neuronal death and Alzheimer's disease.
