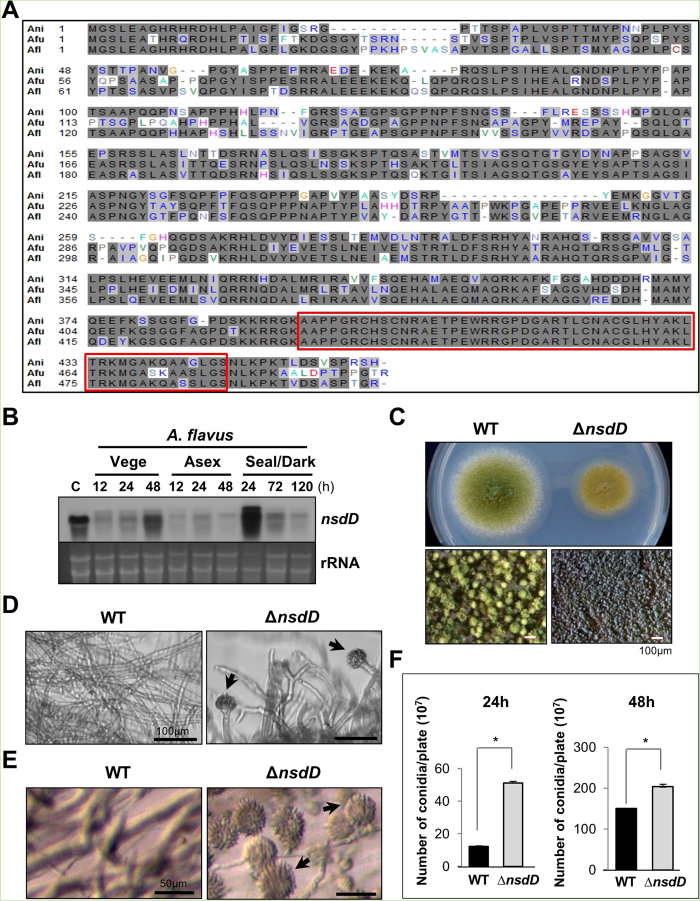
Figure 3. Characterization of nsdD in A. flavus.
(A) Alignment the NsdD proteins of A. fumigatus (Afu3g13870), A. flavus (AFL2T_03635), and A. nidulans (AN3152). The red box indicates the highly conserved region. (B) Levels of nsdD mRNA during the lifecycle of A. flavus WT (NRRL3357). C = Conidia. The time (h) of incubation in liquid submerged culture (Vege), post asexual induction (Asex), and sealed/dark condition (Seal/Dark) is shown. Equal loading of total RNA was confirmed by ethidium bromide staining of rRNA. (C) Phenotypes of A. flavus WT and ΔnsdD (LNJ11) strains point inoculated on solid MMG with 0.1% yeast extract (YE) and incubated at 30 °C for 3 days. Close-up views (lower panel) of the center of individual colonies. (Bar = 100 μm). (D) Cells of A. flavus WT and ΔnsdD strains in liquid submerged culture grown for 28 h. Note the abundant formation of conidiophores in ΔnsdD strain (Bar = 100 μm). Conidiophore is marked by arrowhead. (E) Agar-embedded cells of A. flavus WT and ΔnsdD strains grown on solid MMG for 28 h. Abundant formation of conidiophores in ΔnsdD strain is evident. (Bar = 50 μm). (F) Quantitative analyses of conidiation in A. flavus WT and ΔnsdD strains. About 105 conidia were spread on solid MMG with 0.1% YE, incubated for 24 and 48 h, and the conidia numbers per plate were counted in triplicates (*P < 0.001).