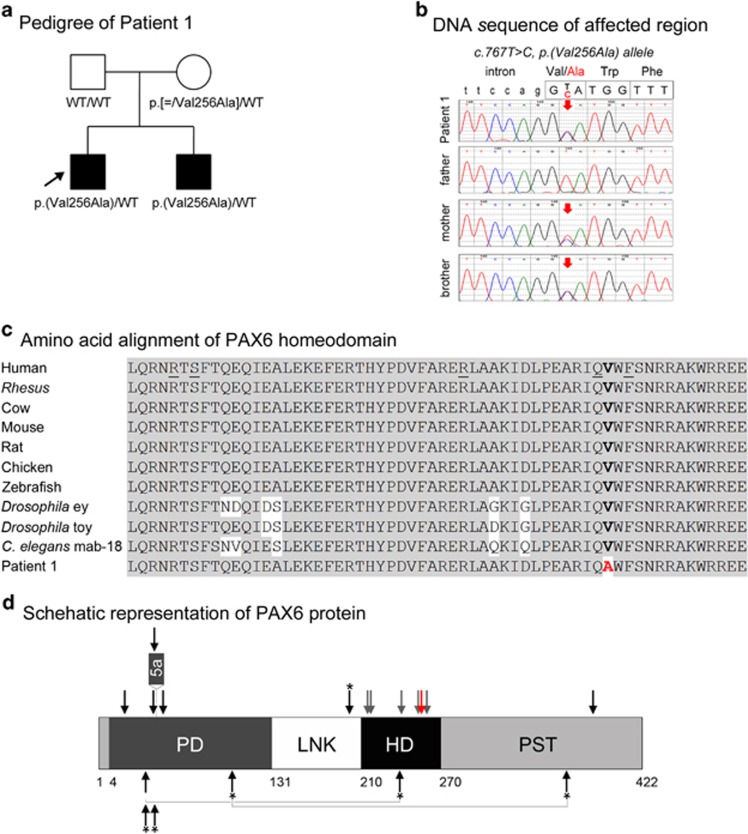
Figure 1.
PAX6 pathogenic variant. (a) Pedigree of Patient 1. Patient 1 (proband) is indicated with an arrow; genotype of each individual is listed below the corresponding symbol. (b) DNA chromatograms for Patient 1 and family members are shown with the position of the c.767T>C, p.(Val256Ala) mutation indicated with red arrows. Please note a smaller ‘C' peak at position c.767 in addition to the wild-type ‘T' in DNA sample of the unaffected mother, suggesting that she is likely to be mosaic for this mutation. (c) Amino-acid alignment of the PAX6 homeodomain from different species. The alignment utilized KALIGN (http://www.ebi.ac.uk/Tools/msa/kalign) and the following reference sequences: AAK95849.1 (human PAX6), AFE78491.1 (Rhesus Pax6), AAI16039.1 (cow Pax6), AAH36957.1 (mouse Pax6), AAI28742.1 (rat Pax6), BAA23004.1 (chicken Pax6), NP_571379.1 and AAC96095.1 (zebrafish pax6a and pax6b, correspondingly; only one HD sequence is shown because it is identical between the two homologs), NM_079889.3 (eyeless (ey); Drosophila melanogaster), AAF59395.4 (twin of eyeless (toy), D. melanogaster), AAA82991.1 (mab-18, Caenorhabditis elegans). The position 256 of the homeodomain is shown in bold and is occupied by a valine residue in all available reference sequences and alanine in Patient 1 and his affected brother. The positions of the previously reported PAX6 mutations affecting the HD are underlined in the human reference sequence. (d) Schematic representation of the PAX6 protein. The positions of PAX6 domains are indicated with different colors and numbers at the bottom of the drawing; PD (paired domain), LNK (linker region), HD (homeodomain), PST (proline–serine–threonine-rich transactivation domain); the position of the protein region encoded by the alternatively spliced exon 5a is also specified. The position of the mutation in Patient 1 is indicated with a red arrow; the positions of published mutations involving the homeodomain are shown with grey arrows; the positions of previously reported A/M mutations are marked with black arrows (for missense) or black arrows with an asterisk (nonsense); dominant (heterozygous) mutations are shown at the top and compound heterozygous mutations (with allelic combinations being connected with a line) on the bottom of the drawing. A full color version of this figure is available at the European Journal of Human Genetics journal online.