Abstract
Stimuli-responsive nanostructures produced with peptide domains from the extracellular matrix offer great opportunities for imaging and drug delivery. Although the individual utility of elastin-like (poly)-peptides and collagen-like peptides in such applications has been demonstrated, the synergistic advantages of combining these motifs in short peptide conjugates have surprisingly not been reported. Here, we introduce the conjugation of a thermoresponsive elastin-like peptide (ELP) with a triple-helix-forming collagen-like peptide (CLP) to yield ELP–CLP conjugates that show a remarkable reduction in the inverse transition temperature of the ELP domain upon formation of the CLP triple helix. The lower transition temperature of the conjugate enables the facile formation of well-defined vesicles at physiological temperature and the unexpected resolubilization of the vesicles at elevated temperatures upon unfolding of the CLP domain. Given the demonstrated ability of CLPs to modify collagens, our results not only provide a simple and versatile avenue for controlling the inverse transition behavior of ELPs, but also suggest future opportunities for these thermoresponsive nanostructures in biologically relevant environments.
In the past few decades, thermoresponsive polymers have been intensely studied to develop new smart materials such as hydrogels, films, and drug nanocarriers. Elastin-like polypeptides (ELPs), in particular, which are derived from the hydrophobic domain of tropoelastin1 and comprise many copies of the pentapeptide repeat Val-Pro-Gly-Xaa-Gly (VPGXG), have also been very widely studied owing to their lower critical solution temperature (LCST)-like behavior. With heating above their inverse transition temperature (Tt), the ELPs collapse into a coacervate phase,2 enabling their use as building blocks for temperature-sensitive smart biomaterials. Many studies have demonstrated the outstanding versatility of the (VPGXG)n consensus repeat for modulating inverse transition temperatures2,3 and in the formation of a range of drug delivery vehicles that can be targeted to tissues and cells via either passive or peptide- and stimuli-responsive mechanisms.4 The inverse temperature transition can also be triggered by cations, such as Ca2+, via functionalization of the ELP with a ligand-binding protein domain.5 While these studies illustrate the utility of the ELPs, essentially all of the ELPs employed have been recombinant, comprising tens or even hundreds of pentapeptide repeats. Short synthetic ELPs (e.g., those with fewer than 10 pentapeptides) have not been used widely for the thermoresponsive fabrication of nanoparticles, owing to their high transition temperatures.3a,b,6 In addition, while many hydrogels and films have been produced from ELPs combined with domains of other structural proteins such as silk and resilin,7 there have been no reports of short ELP-based nanostructures equipped with such domains.
Short synthetic collagen-like peptides (CLPs), similarly, have been employed widely in studies aimed at collagen folding and at development of therapeutic matrices and molecules. CLPs have been shown to mimic the triple helix conformation of native collagen,8 and thus have served as model systems for triple helix structure and the stabilization effect of specific amino acid residues in collagens,9 as well as to mimic collagen fibril formation.10 Additionally, recent studies have illustrated that single-stranded CLPs have a strong propensity to bind native collagen via a strand invasion process.11 The high propensity of CLPs for collagen permits detection of minute quantities of collagen (e.g., 5 ng)12 with substantial promise for staining collagens in human tissues (e.g., skin, cornea, bone,12 and liver13), especially those with high ECM turnover (e.g., prostate tumor xenografts, joints, and articular cartilage14). Despite this widespread use, the utilization of CLPs as domains in responsive nanoparticles has been described in only a very limited number of reports.15
We postulated that the conjugation of short ELPs with CLP domains would offer significant opportunities in the design and application of thermoresponsive nanoparticles, and report here the facile chemical production of these conjugates and their unexpected thermally responsive behavior. The CLP sequence (GPO)4GFOGER(GPO)4GG was employed, owing to the fact that CLPs with eight or more GPO repeats exhibit melting temperatures (Tm) above 37 °C,8a,b,16 which enables formation of stable triple helix at physiological temperature. The peptide sequence GFOGER was employed owing to the fact that it is widely recognized by several kinds of integrins such as α1β2, α2β1, and α11β1.17 It has been reported more recently that the introduction of the GFOGER peptide in a PEG-based hydrogel provides not only a better chondrogenic microenvironment compared with that imparted by the RGD peptide but also enhanced gene expression of type II collagen.18 Based on these investigations, inclusion of the GFOGER domain should facilitate the binding of these materials with cells in future studies. An ELP with the sequence (VPGFG)6 was introduced as the thermoresponsive domain, as it would be expected to have a Tt below 37 °C,3b allowing the conjugate to assemble via collapse of the ELP domain at physiological temperature.
An alkyne-functionalized ELP and azide-functionalized CLP were synthesized via standard Fmoc-based solid-phase peptide synthesis methods (Scheme S1) and purified via reverse-phase high-performance liquid chromatography (HPLC). The purity and expected composition of the peptides were verified by analytical HPLC and electrospray ionization mass spectrometry, respectively (Figures S1–S3). The ELP was then conjugated to the CLP in dimethylformamide via standard copper-catalyzed azide–alkyne cycloaddition methods (Scheme 1); successful synthesis and purification of the conjugates in high yield were verified via gel permeation chromatography, 1H NMR spectroscopy, as well as FT-IR spectroscopy (Figures S4–S8).
Scheme 1.
Chemical Conjugation of ELP and CLP
The ability of the CLP domain to form a stable triple helix at physiological temperature while conjugated to the ELP was probed via circular dichroism (CD) spectroscopy. The CD spectra of ELP–CLP at temperatures ranging from 5 to 80 °C (Figure 1a) show a clear maximum at ca. 225 nm, indicating that the CLP domain is competent to form triple-helical structures after conjugation with ELP. The reduction of the intensity of the peak with increasing temperature (Figure 1b) indicates the expected unfolding of the triple helix upon heating, with the first derivative of the melting curve (after correction for the contribution from the ELP, Figure S9) suggesting a Tm of ca. 57 °C for the ELP–CLP conjugate, which is significantly higher than that of the isolated CLP (ca. 50 °C, Figure S10). Presumably, the collapse of the ELP domain at the elevated temperatures anchors the CLP and stabilizes it against unfolding, similar to our previously reported results for a polymer-conjugated CLP.15b,19 The refolding of the CLP triple helix (Figure 1c,d) is likewise accelerated by the ELP anchoring of the CLP, owing to the increase in the local concentration of the CLP strands.15,19,20 While a rate constant of 1.14 × 107 M−2·s−1 was observed for the refolding reaction of the CLP, the diblock shows a higher rate constant of 4.63 × 107 M−2·s−1.
Figure 1.
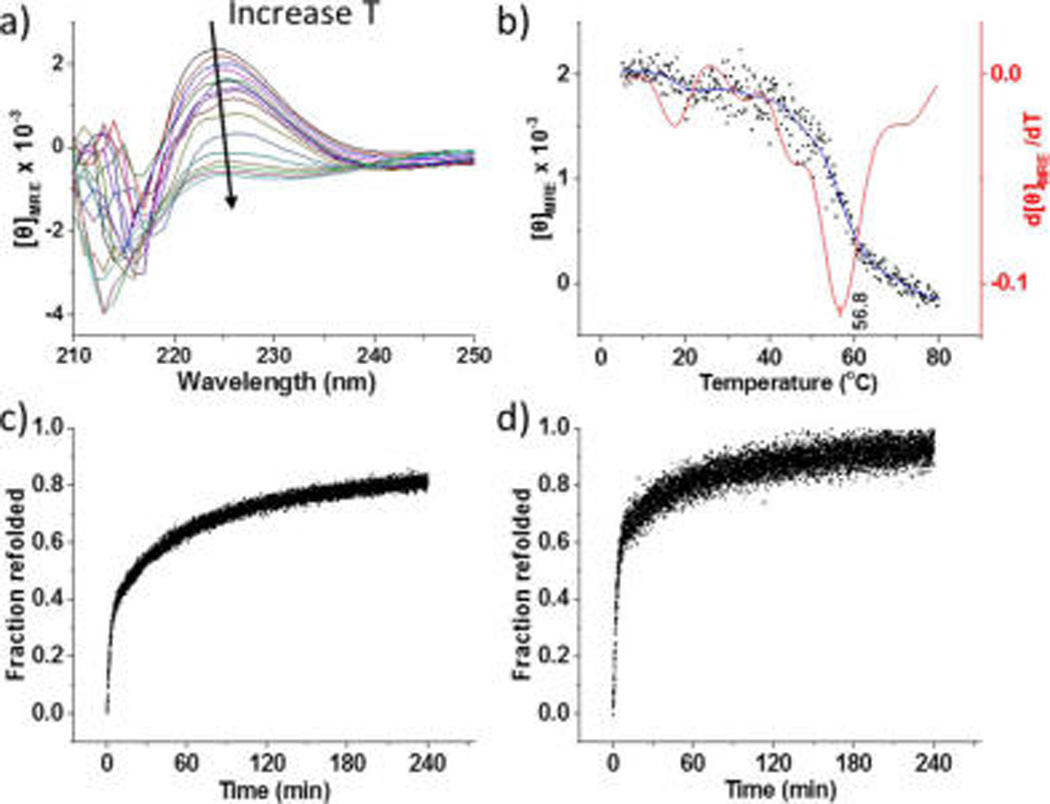
(a) CD spectra showing representative full-wavelength scans for the ELP–CLP conjugate. (b) Thermal unfolding profile for the ELP–CLP conjugate; the first derivative of the unfolding curve with respect to temperature is shown in red. (c) Refolding profile of CLP after quenching from 80 to 5 °C. (d) Refolding profile of the ELP–CLP conjugate after quenching.
The anticipated assembly of ELP–CLP nanostructures at physiologically relevant temperatures was confirmed via dynamic light scattering (DLS, Figure 2). Unexpectedly, the conjugates formed structures with hydrodynamic diameters (Dh) that ranged from approximately 50 to 200 nm at all temperatures between 4 and 65 °C (Figure 2a), with a Dh of approximately 160 nm at 37 °C (Figure 2b). These results are counterintuitive based on the expected increase of the transition temperature of thermoresponsive polymers and ELPs with conjugation to a hydrophilic domain.21 Instead, conjugation of the short ELP to a hydrophilic CLP results in a dramatic reduction of the Tt of the ELP to below 4 °C; the lack of aggregation of the ELP alone (Figure S11) indicates that this reduction exceeds 80 °C.
Figure 2.
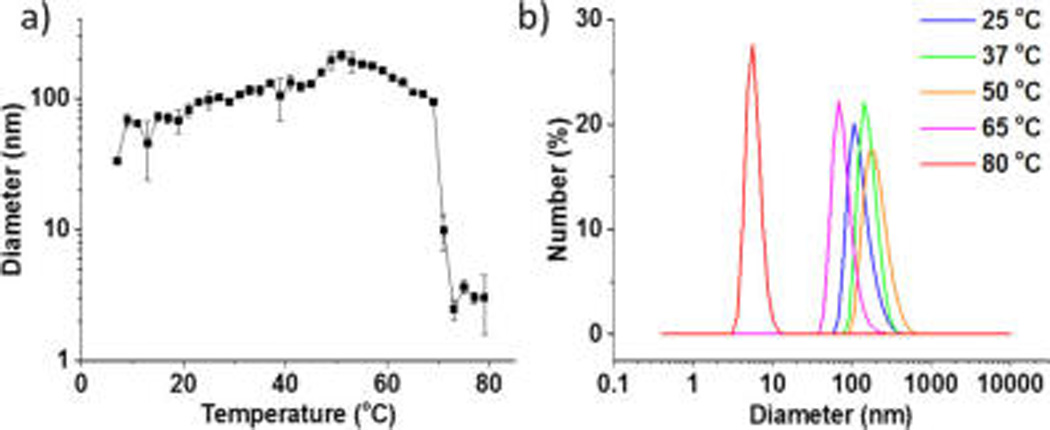
Dynamic light scattering characterization of the assembly of ELP–CLP conjugates. (a) Hydrodynamic diameter of nanostructures as a function of temperature upon heating. (b) Size distributions of ELP– CLP assemblies at select temperatures.
Just as the CLP triple helix is stabilized at high temperature by the anchoring effect of ELP coacervation, the unexpected assembly of the ELP–CLP conjugates at low temperatures is almost certainly attributable to the anchoring effects of the CLP triple helix, which would serve to locally isolate three ELP domains at concentrations approximately 100-fold higher than that of the ELP monomers in solution. Previous investigations have illustrated that ELPs exhibit lower transition temperatures with increasing concentration and length of the (poly)-peptide,3a,b,22 as well as with covalent conjugation of short ELPs as side chains of synthetic graft polymers;23 the colocalization of three ELP chains by the CLP triple helix may thus be expected to show similar trends.
The anchoring of the ELP by the noncovalent formation of CLP triple helix, however, should offer unique and as yet unreported opportunities to reversibly modulate the transition temperatures of the ELP domain and to thus confer dual thermoresponsiveness to the conjugates. Indeed, after an initial increase in the Dh of the ELP–CLP nanoparticles with heating to 50 °C (Figure 2), Dh begins to decrease once the sample is heated above this temperature, which is also approximately the melting temperature of the CLP (Figure 1b). With additional heating to 80 °C, the CLP unfolds completely (Figure 1b), and the nanoparticles become fully solubilized as monomers with an average Dh of only 5.6 nm (Figure 2b). Once the triple helix is unfolded and the ELP is no longer anchored, the Tt of the unfolded ELP–CLP conjugate is above 80 °C, which is consistent with our control results (Figure S11) and with the expected behavior of the ELP with the addition of a hydrophilic CLP domain. This behavior is fully reversible (Figure S12), thus offering a new avenue for controlling the temperature responsiveness of short ELPs.
Transmission electron microscopy (TEM) was conducted to investigate the morphology of the nanostructures formed at 25, 37, 50, 65, and 80 °C (Figure 3a–e, respectively). Consistent with the DLS results, nanoparticles with an average diameter of approximately 80–100 nm were observed at room temperature, and the diameter of these particles increased to 150–250 nm at physiological temperature. Once the sample was heated above the Tm of the collagen domain (50 °C), the nanoparticles showed some changes in morphology and size, with both porosity and apparent monomer (indicated by black arrows) observed at 50 °C (Figure 3c), with increasing porosity and decreasing size when the temperature was raised to 65 °C (Figure 3d). A vesicular structure is suggested for the nanoparticles, although only at the elevated temperatures, perhaps because the PTA stain was capable of diffusing into the porous nanoparticles and thus accumulating at both the exterior and interior surfaces of the vesicles. At 80 °C (Figure 3e), the molecules are soluble and no defined nanostructure was observed.
Figure 3.
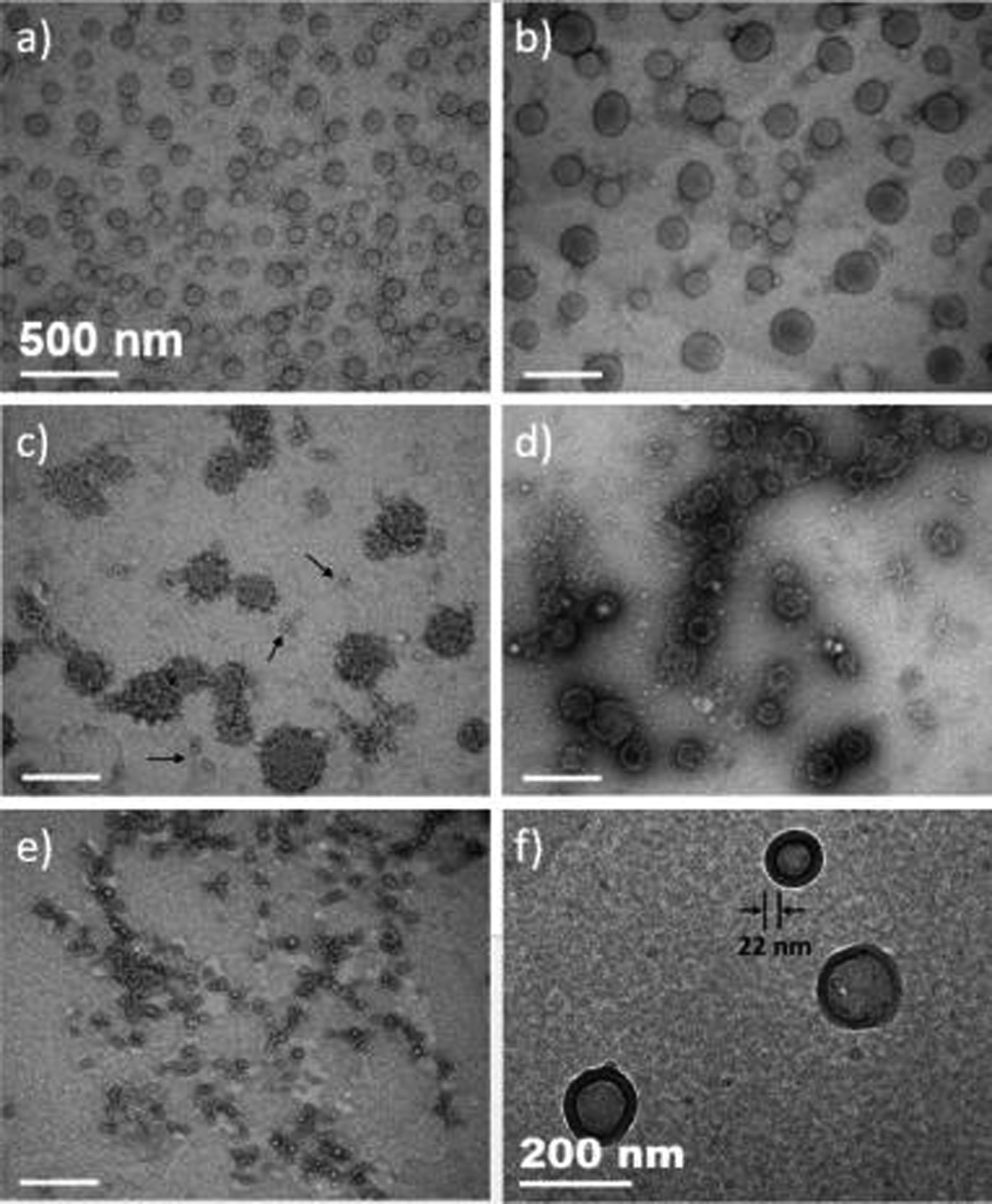
(a–e) TEM images of nanoparticles from ELP–CLP conjugates at various temperatures, after negative staining with phosphotungstic acid: (a) 25, (b) 37, (c) 50, (d) 65, and (e) 80 °C. Scale bars = 500 nm. (f) Cryo-TEM image of nanoparticles of the ELP–CLP diblock at 25 °C. Scale bar = 200 nm.
The vesicular structure of the nanoparticles was further confirmed via cryo-TEM of conjugates incubated at room temperature (Figure 3f); vesicles with a diameter of approximately 100 nm were observed. Image analysis indicates that the thickness of the vesicle walls is 22 ± 2 nm, which is consistent with the presence of two CLP triple helices (9.1 nm each) summed with the approximate Rg of the collapsed ELP domains (3.4 nm).24 The results suggest a bilayer structure of the vesicle walls, with collapsed ELP domains in the center and CLP triple-helical domains at both interior and exterior surfaces (Scheme 2). The presence of a small percentage of unfolded CLP chains is rendered in the schematic, to reflect the unfolding of the CLP domain observed in CD experiments.
Scheme 2.
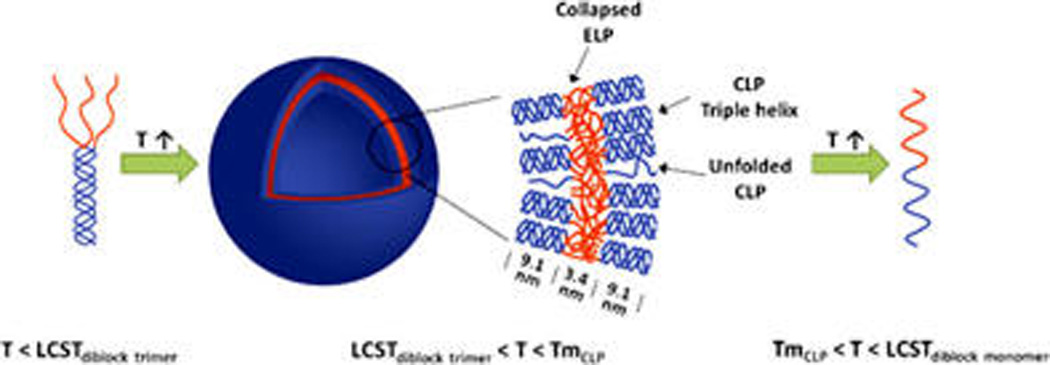
Proposed Assembly/Disassembly and Bilayer Structure of ELP–CLP Vesicles
The thermally induced assembly of ELP block copolypeptides has been a subject of intense investigation over decades. Essentially all previous reports, however, employ ELP-based recombinant polypeptides that mainly form micellar structures, although there are some reports of larger structures and nanostructures that exhibit additional sensitivity to pH and dications.4f,25 There have been very limited reports of thermoresponsive nanostructures that can be assembled from short synthetic ELPs.6,26 Our studies illustrate that this barrier can be overcome by simply anchoring three ELP chains to a collagen triple helix. This not only exploits the reversibility of triple helix formation to modulate the transition temperature of the molecules over a wide range, but should also permit manipulation of the size of the vesicles. In addition, there are few reports of the assembly of thermoresponsive collagen-like peptide containing copolymers,15a and none to our knowledge in which nanovesicles are produced. The likely location of the collagen domain at the exterior surface of the vesicles may serve as a means to localize nanoparticles in collagen-containing tissues, hydrogels, and films.14,27
Simple variations in the relative lengths of the ELP and CLP domains, as well as variations in the sequences of the domains, offer a wide range of options for tailoring the thermoresponsive behavior of these systems. For example, preliminary studies of ELP–CLP conjugates with shorter ELP sequences ((VPGFG)3–5) suggest that the transition temperature of nanoparticle formation and disassembly can be tuned to fall within the physiological range for the (VPGFG)5-CLP (Figure S13). The large size and polydispersity of the aggregates, however, suggest that the hydrophobic interactions of the shorter (VPGFG)5 are insufficient to form well-defined nanoparticles. Changes to the stability of the CLP block, when balanced with the hydrophobicity of the ELP domain, could also be employed to impart triggered assembly/disassembly under select conditions. The prospects are promising for these approaches in drug delivery, imaging, and materials modification.
Supplementary Material
Acknowledgments
This work was funded in part by the National Science Foundation (DMR 0907478 to K.L.K.) and the National Center for Research Resources (NCRR), a component of the National Institutes of Health (1 P30 GM103519 and 1 P30 GM110758-01 for instrument resources). We thank Chaoying Ni, Fei Deng, Yingchao Chen, and Darrin Pochan at the University of Delaware for advice and assistance with TEM, and Rachel Letteri at University of Massachusetts Amherst for conducting GPC (supported by the Materials Research Facilities Network (MRFN) of the UMass MRSEC on Polymers (DMR-0820506)).
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Experimental details; peptide and diblock synthesis; diblock characterization; and CD and DLS data, including Scheme S1 and Figures S1–S13 (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Foster JA, Bruenger E, Gray WR, Sandberg LB. J. Biol. Chem. 1973;248:2876–2879. [PubMed] [Google Scholar]
- 2.Urry DW. J. Phys. Chem. B. 1997;101:11007–11028. [Google Scholar]
- 3.(a) McDaniel JR, Radford DC, Chilkoti A. Biomacromolecules. 2013;14:2866–2872. doi: 10.1021/bm4007166. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nuhn H, Klok HA. Biomacromolecules. 2008;9:2755–2763. doi: 10.1021/bm800784y. [DOI] [PubMed] [Google Scholar]; (c) Quiroz FG, Chilkoti A. Nat. Mater. 2015;14:1164–1171. doi: 10.1038/nmat4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Martin L, Castro E, Ribeiro A, Alonso M, Rodriguez-Cabello JC. Biomacromolecules. 2012;13:293–298. doi: 10.1021/bm201436y. [DOI] [PubMed] [Google Scholar]; (b) Park WM, Champion JA. J. Am. Chem. Soc. 2014;136:17906–17909. doi: 10.1021/ja5090157. [DOI] [PubMed] [Google Scholar]; (c) Garcia-Arevalo C, Bermejo-Martin JF, Rico L, Iglesias V, Martin L, Rodriguez-Cabello JC, Arias FJ. Mol. Pharmaceutics. 2013;10:586–597. doi: 10.1021/mp300325v. [DOI] [PubMed] [Google Scholar]; (d) Fujita Y, Mie M, Kobatake E. Biomaterials. 2009;30:3450–3457. doi: 10.1016/j.biomaterials.2009.03.012. [DOI] [PubMed] [Google Scholar]; (e) Shi P, Gustafson JA, MacKay JA. Int. J. Nanomed. 2014;9:1617–1626. doi: 10.2147/IJN.S53886. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Callahan DJ, Liu WE, Li XH, Dreher MR, Hassouneh W, Kim M, Marszalek P, Chilkoti A. Nano Lett. 2012;12:2165–2170. doi: 10.1021/nl300630c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Dreher MR, Simnick AJ, Fischer K, Smith RJ, Patel A, Schmidt M, Chilkoti A. J. Am. Chem. Soc. 2008;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim B, Chilkoti A. J. Am. Chem. Soc. 2008;130:17867–17873. doi: 10.1021/ja8059057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Eldijk MB, Wang JCY, Minten IJ, Li CL, Zlotnick A, Nolte RJM, Cornelissen J, van Hest JCM. J. Am. Chem. Soc. 2012;134:18506–18509. doi: 10.1021/ja308132z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Haghpanah JS, Tu R, Da Silva S, Yan D, Mueller S, Weder C, Foster EJ, Sacui I, Gilman JW, Montclare JK. Biomacromolecules. 2013;14:4360–4367. doi: 10.1021/bm401304w. [DOI] [PubMed] [Google Scholar]; (b) Liu JC, Tirrell DA. Biomacromolecules. 2008;9:2984–2988. doi: 10.1021/bm800469j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Dinerman AA, Cappello J, El-Sayed M, Hoag SW, Ghandehari H. Macromol. Biosci. 2010;10:1235–1247. doi: 10.1002/mabi.201000061. [DOI] [PubMed] [Google Scholar]; (d) Nowatzki PJ, Tirrell DA. Biomaterials. 2004;25:1261–1267. doi: 10.1016/s0142-9612(03)00635-5. [DOI] [PubMed] [Google Scholar]; (e) Gustafson JA, Price RA, Greish K, Cappello J, Ghandehari H. Mol. Pharmaceutics. 2010;7:1050–1056. doi: 10.1021/mp100161u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Gustafson JA, Ghandehari H. Adv. Drug Delivery Rev. 2010;62:1509–1523. doi: 10.1016/j.addr.2010.04.006. [DOI] [PubMed] [Google Scholar]; (g) Trabbic-Carlson K, Setton LA, Chilkoti A. Biomacromolecules. 2003;4:572–580. doi: 10.1021/bm025671z. [DOI] [PubMed] [Google Scholar]; (h) McHale MK, Setton LA, Chilkoti A. Tissue Eng. 2005;11:1768–1779. doi: 10.1089/ten.2005.11.1768. [DOI] [PubMed] [Google Scholar]; (i) Bracalello A, Santopietro V, Vassalli M, Marletta G, Del Gaudio R, Bochicchio B, Pepe A. Biomacromolecules. 2011;12:2957–2965. doi: 10.1021/bm2005388. [DOI] [PubMed] [Google Scholar]
- 8.(a) Luo TZ, Kiick KL. Eur. Polym. J. 2013;49:2998–3009. doi: 10.1016/j.eurpolymj.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yu SM, Li Y, Kim D. Soft Matter. 2011;7:7927–7938. doi: 10.1039/C1SM05329A. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Shoulders MD, Raines RT. Annual Reviews in Biochemistry. Vol. 78. Palo Alto, CA: Annual Reviews; 2009. Collagen Structure and Stability; pp. 929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Fallas JA, Dong JH, Tao YZJ, Hartgerink JD. J. Biol. Chem. 2012;287:8039–8047. doi: 10.1074/jbc.M111.296574. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Persikov AV, Ramshaw JAM, Brodsky B. J. Biol. Chem. 2005;280:19343–19349. doi: 10.1074/jbc.M501657200. [DOI] [PubMed] [Google Scholar]; (c) He LR, Theato P. Eur. Polym. J. 2013;49:2986–2997. [Google Scholar]
- 10.(a) O’Leary LER, Fallas JA, Bakota EL, Kang MK, Hartgerink JD. Nat. Chem. 2011;3:821–828. doi: 10.1038/nchem.1123. [DOI] [PubMed] [Google Scholar]; (b) Rele S, Song YH, Apkarian RP, Qu Z, Conticello VP, Chaikof EL. J. Am. Chem. Soc. 2007;129:14780–14787. doi: 10.1021/ja0758990. [DOI] [PubMed] [Google Scholar]; (c) Krishna OD, Kiick KL. Biomacromolecules. 2009;10:2626–2631. doi: 10.1021/bm900551c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang AY, Mo X, Chen CS, Yu SM. J. Am. Chem. Soc. 2005;127:4130–4131. doi: 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Ho D, Meng H, Chan TR, An B, Yu H, Brodsky B, Jun AS, Yu SM. Bioconjugate Chem. 2013;24:9–16. doi: 10.1021/bc3005842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang AY, Foss CA, Leong S, Mo X, Pomper MG, Yu SM. Biomacromolecules. 2008;9:1755–1763. doi: 10.1021/bm701378k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Foss CA, Summerfield DD, Doyle JJ, Torok CM, Dietz HC, Pomper MG, Yu SM. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14767–14772. doi: 10.1073/pnas.1209721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Luo JN, Tong YW. ACS Nano. 2011;5:7739–7747. doi: 10.1021/nn202822f. [DOI] [PubMed] [Google Scholar]; (b) Krishna OD, Wiss KT, Luo TZ, Pochan DJ, Theato P, Kiick KL. Soft Matter. 2012;8:3832–3840. doi: 10.1039/C2SM07025A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brodsky B, Persikov AV. Molecular structure of the collagen triple helix. In: Parry DAD, Squire JM, editors. Fibrous Proteins: Coiled-Coils, Collagen and Elastomers. Vol. 70. San Diego, CA: Elsevier Academic Press Inc.; 2005. p. 301–+. [DOI] [PubMed] [Google Scholar]
- 17.(a) Knight CG, Morton LF, Peachey AR, Tuckwell DS, Farndale RW, Barnes MJ. J. Biol. Chem. 2000;275:35–40. doi: 10.1074/jbc.275.1.35. [DOI] [PubMed] [Google Scholar]; (b) Zhang WM, Kapyla J, Puranen JS, Knight CG, Tiger CF, Pentikainen OT, Johnson MS, Farndale RW, Heino J, Gullberg D. J. Biol. Chem. 2003;278:7270–7277. doi: 10.1074/jbc.M210313200. [DOI] [PubMed] [Google Scholar]; (c) Reyes CD, Garcia AJ. J. Biomed. Mater. Res. 2003;65A:511–523. doi: 10.1002/jbm.a.10550. [DOI] [PubMed] [Google Scholar]
- 18.Mhanna R, Ozturk E, Vallmajo-Martin Q, Millan C, Muller M, Zenobi-Wong M. Tissue Eng., Part A. 2014;20:1165–1174. doi: 10.1089/ten.tea.2013.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo TZ, He LR, Theato P, Kiick KL. Macromol. Biosci. 2015;15:111–123. doi: 10.1002/mabi.201400358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu YC, Berndt P, Tirrell M, Fields GB. J. Am. Chem. Soc. 1996;118:12515–12520. [Google Scholar]
- 21.(a) Isoda K, Kanayama N, Miyamoto D, Takarada T, Maeda M. React. Funct. Polym. 2011;71:367–371. [Google Scholar]; (b) Zhao CW, Zhuang XL, He CL, Chen XS, Jing XB. Macromol. Rapid Commun. 2008;29:1810–1816. [Google Scholar]; (c) Chilkoti A, Dreher MR, Meyer DE. Adv. Drug Delivery Rev. 2002;54:1093–1111. doi: 10.1016/s0169-409x(02)00060-1. [DOI] [PubMed] [Google Scholar]; (d) MacKay JA, Callahan DJ, FitzGerald KN, Chilkoti A. Biomacromolecules. 2010;11:2873–2879. doi: 10.1021/bm100571j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer DE, Chilkoti A. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 23.(a) Fernandez-Trillo F, Dureault A, Bayley JPM, van Hest JCM, Thies JC, Michon T, Weberskirch R, Cameron NR. Macromolecules. 2007;40:6094–6099. [Google Scholar]; (b) Conrad RM, Grubbs RH. Angew. Chem., Int. Ed. 2009;48:8328–8330. doi: 10.1002/anie.200903888. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ayres L, Koch K, Adams P, van Hest JCM. Macromolecules. 2005;38:1699–1704. [Google Scholar]
- 24.(a) Ainavarapu RK, Brujic J, Huang HH, Wiita AP, Lu H, Li LW, Walther KA, Carrion-Vazquez M, Li HB, Fernandez JM. Biophys. J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Stirnemann G, Giganti D, Fernandez JM, Berne BJ. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3847–3852. doi: 10.1073/pnas.1300596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) Kim WY, Xiao JT, Chaikof EL. Langmuir. 2011;27:14329–14334. doi: 10.1021/la202864x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kim W, Thevenot J, Ibarboure E, Lecommandoux S, Chaikof EL. Angew. Chem., Int. Ed. 2010;49:4257–4260. doi: 10.1002/anie.201001356. [DOI] [PubMed] [Google Scholar]; (c) Dash BC, Mahor S, Carroll O, Mathew A, Wang WX, Woodhouse KA, Pandit A. J. Controlled Release. 2011;152:382–392. doi: 10.1016/j.jconrel.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 26.(a) Pechar M, Brus J, Kostka L, Konak C, Urbanova M, Slouf M. Macromol. Biosci. 2007;7:56–69. doi: 10.1002/mabi.200600196. [DOI] [PubMed] [Google Scholar]; (b) Dai M, Haghpanah J, Singh N, Roth EW, Liang A, Tu RS, Montclare JK. Biomacromolecules. 2011;12:4240–4246. doi: 10.1021/bm201083d. [DOI] [PubMed] [Google Scholar]
- 27.(a) Mo X, An YJ, Yun CS, Yu SM. Angew. Chem., Int. Ed. 2006;45:2267–2270. doi: 10.1002/anie.200504529. [DOI] [PubMed] [Google Scholar]; (b) Urello MA, Kiick KL, Sullivan MO. J. Mater. Chem. B. 2014;2:8174–8185. doi: 10.1039/c4tb01435a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



