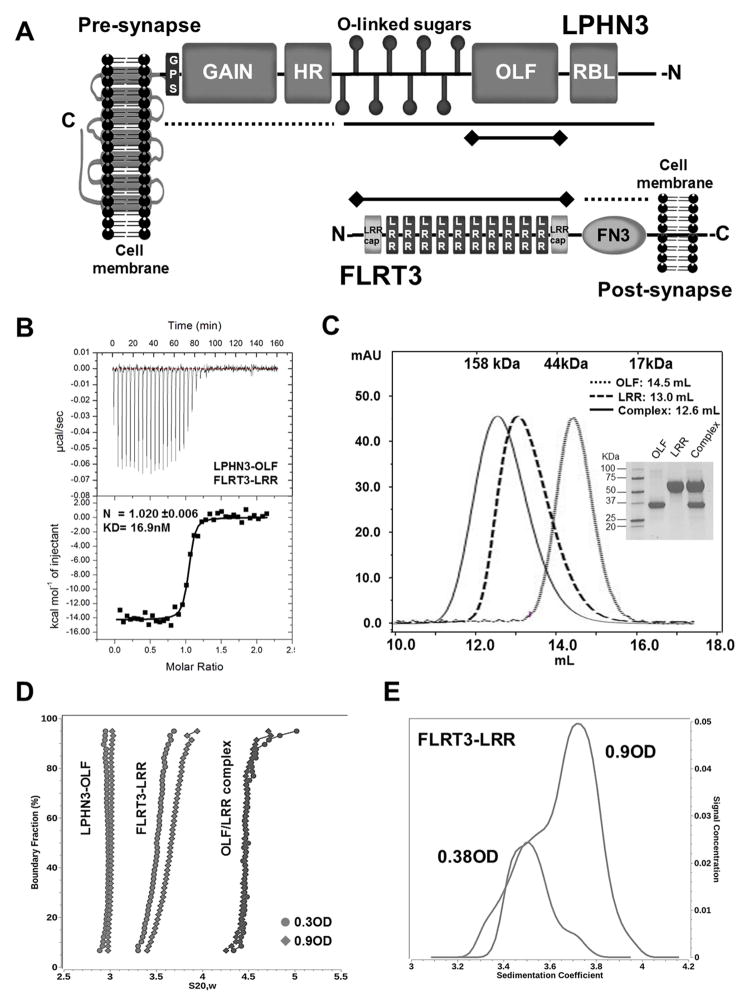
Figure 1. Characterization of LPHN3-OLF and FLRT3-LRR in solution.
(A), Schematic representation of LPHN3 and FLRT3 domain architecture. N- and C- designate N- and C-termini of the proteins. Dotted lines indicate constructs that did not bind, solid lines indicate constructs that bound and solid lines with terminal diamonds indicate constructs used in B. (B), ITC of LPHN3-OLF injected at 100μM into the cell containing the FLRT3-LRR domain at 10μM. (C), SEC of the purified LPHN3-OLF, FLRT3-LRR and their complex using a Superdex 200-10/300GL Column. Top margin shows the column calibration: 1, γ-globulin, 158KDa; 2, Ovalbumin, 44KDa; 3, myoglobin, 17KDa. OLF eluted with an apparent MW of ~30KDa; LRR at ~ 71KDa; the complex at ~90KDa as expected for a 1:1 stoichiometry. Inset, Coomassie blue staining SDS-PAGE of the purified proteins. (D), Sedimentation velocity of LPHN3-OLF, FLRT3-LRR and their complex, at two concentrations. Overlay of the diffusion corrected integral sedimentation coefficient distributions from the van Holde - Weischet analysis shows a single species with homogeneous distribution for LPHN3-OLF and the complex but significant self-association for FLRT-LRR. The frictional ratio of the monomer suggested a slightly non-globular shape and was determined to be 1.53 (1.48 and 1.58 with 95% confidence intervals). The dimer frictional ratio was found to be more elongated at 1.76 (1.74 and 1.78 with 95% confidence intervals). (E), The van Holde - Weischet analysis of the FLRT3-LRR concentrations reveal two distinct but overlapping species indicating a clear monomer-dimer equilibrium.