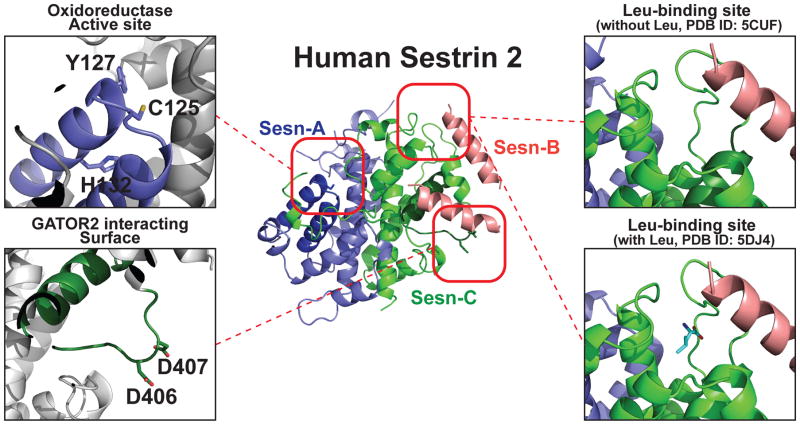
Figure 1. Structure of hSesn2 with highlighted functional domains.
Ribbon diagram of full-length hSesn2 (PDB ID: 5CUF [21]). Sesn-A, Sesn-B and Sesn-C domains, which were originally predicted through a phylogenetic analysis [73], are represented in blue, pink and green, respectively. The oxidoreductase active site of Sesn-A is magnified in the top left, and the catalytic cysteine (C125) and conserved residues of the proton relay system (Y127 and H132) are indicated. These residues were found to be critical for the antioxidant function of hSesn2. The GATOR2-interacting surface of the Sesn-C domain is magnified on the bottom left with the critical residues (D406 and D407; the ‘DD motif’) labeled. This DD motif is required for the interaction between hSesn2 and GATOR2, and for subsequent control of AMPK and mTORC1 signaling. Finally, the leucine-binding site of the Sesn-C domain is magnified on the right. The top representation is the crystallized structure without added leucine (PDB ID: 5CUF [21]), and the bottom representation is the structure with leucine (PDB ID: 5DJ4 [22]). No significant conformational change was observed between these two structures. Illustrations of the protein structure were generated with PYMOL (Delano Scientific, LLC).