Fig. 4.
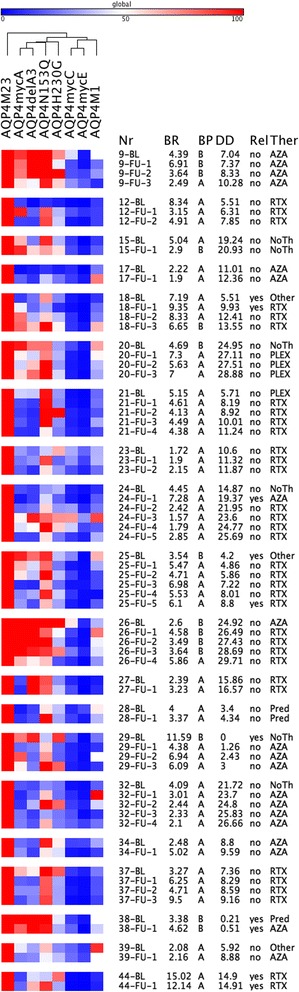
Heatmap of serum AQP4-antibody levels against AQP4-M23, AQP4-M1 and AQP4-M23 mutants (columns) in baseline and follow-up samples of 20 NMOSD patients with follow-up samples. Rows are individual samples with patient IDs (Nr), flow cytometry AQP4-M23 binding ratios (BR), AQP4-IgG binding patterns (BP), disease duration in years (DD), presence of acute relapses (Rel), and therapies (Ther) shown at the right side. Data are shown as percent binding of AQP4-M23. Values range from blue (0 %) to white (50 %) to red (100 %). Columns were clustered according to their Pearson’s correlation coefficients. Two major antibody binding patterns were identified, a loop A-dependent pattern A and an independent pattern B. The heatmap was generated using GENE-E matrix visualization and analysis software (http://www.broadinstitute.org/cancer/software/GENE-E/index.html). AZA azathioprine, NoTh no therapy, other other immunsuppressive therapies, PLEX plasma exchange, Pred _ corticosteroids, RTX rituximab
