Abstract
The basal-like breast cancer (BLBC) subtype accounts for a disproportionately high percentage of overall breast cancer mortality. The current therapeutic options for BLBC need improvement; hence, elucidating signaling pathways that drive BLBC growth may identify novel targets for the development of effective therapies. Rho GTPases have previously been implicated in promoting tumor cell proliferation and metastasis. These proteins are inactivated by Rho-selective GTPase-activating proteins (RhoGAPs), which have generally been presumed to act as tumor suppressors. Surprisingly, RNA-Seq analysis of the Rho GTPase signaling transcriptome revealed high expression of several RhoGAP genes in BLBC tumors, raising the possibility that these genes may be oncogenic. To evaluate this, we examined the roles of two of these RhoGAPs, ArhGAP11A (also known as MP-GAP) and RacGAP1 (also known as MgcRacGAP), in promoting BLBC. Both proteins were highly expressed in human BLBC cell lines, and knockdown of either gene resulted in significant defects in the proliferation of these cells. Knockdown of ArhGAP11A caused CDKN1B/p27-mediated arrest in the G1 phase of the cell cycle, whereas depletion of RacGAP1 inhibited growth through the combined effects of cytokinesis failure, CDKN1A/p21-mediated RB1 inhibition, and the onset of senescence. Random migration was suppressed or enhanced by the knockdown of ArhGAP11A or RacGAP1, respectively. Cell spreading and levels of GTP-bound RhoA were increased upon depletion of either GAP. We have established that, via the suppression of RhoA, ArhGAP11A and RacGAP1 are both critical drivers of BLBC growth, and propose that RhoGAPs can act as oncogenes in cancer.
Keywords: ArhGAP11A, RacGAP1, basal-like breast cancer, RhoA, RhoGAP
Introduction
Human breast tumors can be classified into one of five major subtypes (luminal A, luminal B, HER2-enriched, basal-like, and normal breast-like) based upon global gene expression analyses (1-3). The basal-like subgroup is responsible for a disproportionately high percentage of overall breast cancer recurrence and death, and accounts for approximately 80% of the ‘triple-negative’ breast cancers (4,5), for which the current standard of care is limited to conventional cytotoxic chemotherapy (6).
Rho family small GTPases (e.g., RhoA, Rac1, and Cdc42) are intracellular signaling molecules belonging to the Ras superfamily (7,8). When GTP-bound, these proteins are active and capable of signaling to a diverse array of downstream effectors, through which they regulate cellular responses such as cell cycle progression, cytokinesis, survival, migration, and polarity. Rho GTPases have been strongly implicated in tumorigenesis (9), yet, until recently, these proteins were rarely identified as being mutated in cancer. Instead, dysregulated activity of Rho GTPases in cancer more commonly arises through their aberrant expression and/or activation.
Rho GTPase activity is regulated by Rho-selective guanine nucleotide exchange factors (RhoGEFs), GTPase-activating proteins (RhoGAPs), and guanine nucleotide dissociation inhibitors (RhoGDIs). In the context of cancer, RhoGEFs, which promote the formation of the active GTP-bound state of Rho GTPases by catalyzing the exchange of GDP for GTP (10), are thought to drive tumor growth when overexpressed or aberrantly activated. For example, we recently showed that the RhoGEF PREX1 is overexpressed in melanoma and that PREX1-deficient mice have impaired metastatic tumor growth (11). In contrast, RhoGAPs, which stimulate the intrinsic GTPase activity of Rho proteins and return them to an inactive, GDP-bound state (12), are generally presumed to inhibit tumorigenesis (13). For example, the RhoGAP DLC1 is commonly lost in cancer through promoter methylation, genomic deletion, or enhanced protein degradation (14,15). Hence, the prevailing dogma in the field is that Rho GTPases and RhoGEFs are oncogenes in cancer, whereas RhoGAPs are tumor suppressors.
Recent evidence has begun to challenge this notion, however. Genomic sequencing has revealed frequent somatic mutations of RhoA in peripheral T cell lymphomas (PTCLs) (16-18) and in diffuse-type gastric carcinomas (19-21). Surprisingly, unlike Rac1, which has gain-of-function mutations in melanoma (22), hotspot mutations in RhoA were identified at sites consistent with loss-of-function. In PTCL, the predominant mutation was G17V, which abolishes GTP-binding and causes RhoA to act as a dominant-negative inhibitor of RhoGEF activity (16-18). Diffuse-type gastric cancers exhibited mutations in the effector binding domain of RhoA, most commonly Y42C (19-21), which prevents binding to the Rho effector PKN1 (23). In addition to these mutational analyses, another recent study has shown that colorectal cancer growth is accelerated in the presence of dominant-negative (T19N) RhoA (24). Taken together, and contrary to the existing paradigm, this emerging evidence suggests that, in certain cancers, wild type RhoA may act as a tumor suppressor.
In this study, we analyzed RNA-Seq data of human breast tumors to identify Rho GTPases or GTPase regulators that are aberrantly expressed in breast cancer. To our surprise, several RhoGAP genes exhibited high expression in the basal-like subtype. Focusing on two of these genes, ARHGAP11A and RACGAP1, we characterized their role(s) in promoting basal-like breast cancer (BLBC). In basal-like cell lines, both GAPs were required for proliferation. Suppression of ArhGAP11A (also known as MP-GAP) expression caused cells to undergo cell cycle arrest, whereas RacGAP1 (also known as MgcRacGAP)-depleted cells failed to grow as a result of the combined effect of cytokinesis failure and the onset of senescence. We propose that ArhGAP11A and RacGAP1 act as oncoproteins in BLBC and that GAP-mediated inhibition of RhoA activity is a pro-proliferative mechanism, consistent with the emerging view of RhoA as a tumor suppressor.
Materials and Methods
Cell Culture
Cells were cultured at 37°C in a humidified/5% CO2 atmosphere. SUM149 and SUM159 cells (Asterand) were maintained in Ham's F-12 medium (Gibco), 10 mM HEPES, 5 μg/ml insulin, 1 μg/ml hydrocortisone, and 5% FBS. HMLE cells (provided by Robert Weinberg, MIT) were cultured in mammary epithelial cell growth medium (Lonza). BT474, HCC1937, HEK293T, MCF10A, MCF7, MDA-MB-231, MDA-MB-468, and T47D cells were purchased from American Type Culture Collection (ATCC) and maintained according to ATCC instructions. All cell lines were obtained between 2003 and 2015 and passaged for fewer than 6 months after receipt/resuscitation from cell banks. For apoptosis experiments, SUM149 or HCC1937 cells were treated with 1 μM staurosporine for 4 or 6 h, respectively.
Vectors and lentivirus preparation
Target short hairpin RNA (shRNA) sequences (in pLKO.1 vectors) were as follows: non-silencing (NS) control 5’-CAACAAGATGAAGAGCACCAA-3’, ArhGAP11A sh3 5’-CTGGTGTCAATAGATATGAAA-3’, ArhGAP11A sh5 5’-CCTTCTATTACACCTCAAGAA-3’, RacGAP1 sh1 5’-CAGGTGGATGTAGAGATCAAA-3’, and RacGAP1 sh2 5’-CTAGGACGACAAGGCAACTTT-3’. A cDNA clone containing the ORF of ARHGAP11A (Genbank accession NM_014783) was generated by subcloning bp 2204-3794 of exon 12 onto the 3’ end of IMAGE clone 5502381 (Genbank accession BC063444, Center for Cancer Systems Biology), using an engineered XhoI site and partial overlapping primers to excise intervening sequences. Full-length RACGAP1 cDNA was from the Center for Cancer Systems Biology (IMAGE clone 5583315, Genbank accession BC032754). ArhGAP11A, RacGAP1, and KRAS4B(G12V) cDNAs were subcloned into the pCDH-HA lentiviral vector (System Biosciences). Lentivirus particles were produced by transfecting HEK293T cells with target vectors and the ViraPower lentiviral packaging system (ThermoFisher Scientific). For lentiviral transduction, cells were incubated with virus for 14 h, passaged 48 h after initial transduction, and then used in experiments after puromycin selection. We have previously described the pCMV-Myc-RhoA Q63L, pCMV-Myc-Rac1 Q61L, and pCMV-Myc-Cdc42 Q61L vectors (25).
Immunoblot analyses
Antibodies recognizing the following proteins were used: ArhGAP11A (Abcam, ab113261, 1:1,000), RacGAP1 (Abnova, H00029127-M01, 1:1,000), Rac1 (BD Biosciences, 610650, 1:500), HA epitope tag (BioLegend, 901513, 1:500), cyclin D1 (Cell Signaling, 2922, 1:1,000), MAPK1/3 (ERK; Cell Signaling, 9102, 1:1,000), p21 (Cell Signaling, 2947, 1:1,000), p27 (Cell Signaling, 2552, 1:1,000), PARP1 (Cell Signaling, 9542, 1:3,000), phospho-ERK (Cell Signaling, 4370, 1:1,000), phospho-RB1 (Cell Signaling, 9308, 1:1,000), phospho-RPS6KA1 (RSK; Cell Signaling, 9344, 1:1,000), RB1 (Cell Signaling, 9309, 1:1,000), RhoA (Cell Signaling, 2117, 1:1,000), actin (EMD Millipore, MAB1501, 1:10,000), Cdc42 (Santa Cruz, sc-87, 1:200), cyclin E1 (Santa Cruz, sc-247, 1:500), and TP53 (Santa Cruz, sc-6243, 1:5,000). Densitometric quantification of blots was performed using ImageJ software (NIH).
Growth assays
For anchorage-dependent clonogenic growth assays, 104 cells/well were grown in 6-well plates in the presence or absence of 10 μM of the ROCK1/2 inhibitor Y-27632 (EMD Millipore, 688000) for 7 (SUM149) or 10 (HCC1937) days, then stained with 0.2% crystal violet in 4% formaldehyde for 20 min. For MTT viability assays, 1,000 cells/well (500 cells/well for SUM149) were grown in 96 well plates for up to 14 days then stained with 0.3 mg/ml MTT for 3 h. After solubilizing in dimethyl sulfoxide, A550 was recorded using a BioTek Synergy 2 plate reader. MCF10A acinar formation assays were performed as previously described (26). For fluorescent microscopy, acini were fixed after 12 days then stained with Alexa Fluor 568 phalloidin (ThermoFisher Scientific, A12380, 1:250) and Hoechst 33342 (ThermoFisher Scientific, H3570, 1:10,000). Images were taken using a Zeiss Axiovert 200M microscope (10× objective), Hamamatsu ORCA-ER camera, and Axiovision software. Acinar perimeter and area were determined using ImageJ software (NIH).
Cell cycle analysis
Cells were fixed in 70% ethanol for at least 30 min, stained with 50 μg/ml propidium iodide in PBS plus 100 μg/ml RNase for 15 min at 37°C, then analyzed for DNA content using a CyAn ADP flow cytometer and Summit software (Beckman Coulter).
Senescence
Senescence-associated β-galactosidase was detected 7 days post-plating using a staining kit (Cell Signaling, 9860), according to the manufacturer's instructions. Images were taken using a Nikon Eclipse TS100 microscope (20× objective) and Apple iPhone 6 camera.
Spreading assays and fluorescence microscopy
Cells were suspended in Ham's F-12 media, 10 mM HEPES, 5 μg/ml insulin, 1 μg/ml hydrocortisone, and 0.5% fatty acid-free bovine serum albumin, rotated end-over-end at 37°C for 1 h, then plated on 10 μg/ml fibronectin-coated coverslips for 2 h. For fluorescent microscopy, cells were fixed and then stained with Alexa Fluor 488 phalloidin (ThermoFisher Scientific, A12379, 1:500) and Hoechst 33342 (ThermoFisher Scientific, H3570, 1:10,000). Images were taken using a Zeiss Axiovert 200M microscope (63× objective), Hamamatsu ORCA-ER camera, and Axiovision software. Cell area was determined using ImageJ software (NIH); multinucleated cells were excluded from the spreading analysis.
Random migration assay
Cells (5 × 104/well) were plated on 10 μg/ml fibronectin in 35 mm MatTek dishes and imaged at 37°C, 5% CO2 every 10 min for 24 h using an Olympus VivaView system (10× objective). Random migration was analyzed using the ImageJ software (NIH) Manual Tracking plugin, and the Ibidi Chemotaxis and Migration Tool.
Rho GTPase pulldowns
GTP-bound RhoA, Rac1, and Cdc42 levels were measured as we have previously described (27).
Results
ArhGAP11A and RacGAP1 are highly expressed in BLBC
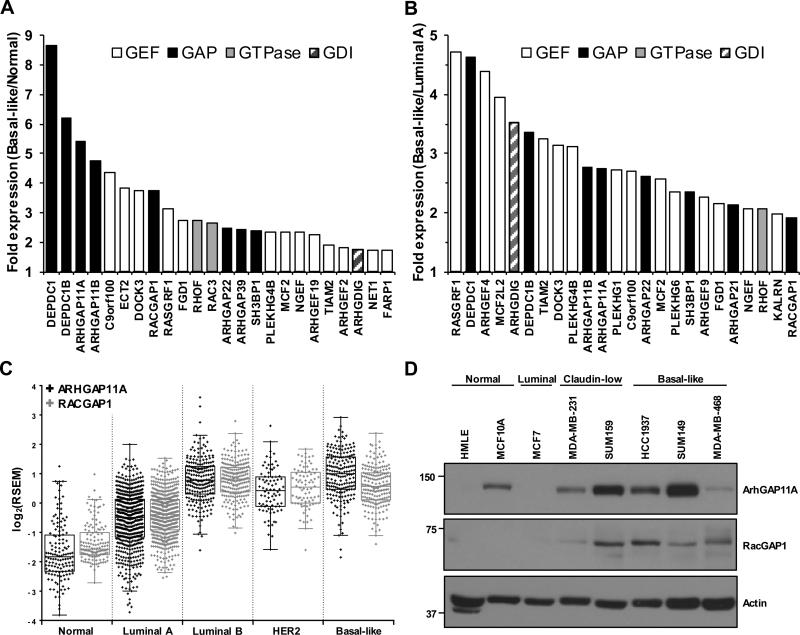
To identify components of Rho GTPase signaling networks that are upregulated in BLBC, we analyzed RNA-Seq data, coming from 1,201 human breast tumors as part of The Cancer Genome Atlas (TCGA) Project (28), for the expression of the 20 Rho GTPase, 79 RhoGEF, 64 RhoGAP, and three RhoGDI genes across different breast cancer subtypes. Strikingly, a number of genes encoding RhoGAPs were found to be highly expressed in tumors of the basal-like subtype relative to normal-like (Fig. 1A) or luminal A (Fig. 1B) tumors, which have a better prognosis. This was surprising, given that RhoGAPs have been generally presumed to act as tumor suppressors. To explore the possibility that RhoGAP genes may in fact play oncogenic roles in BLBC, we chose to focus on two GAP genes that were among the most highly upregulated Rho GTPase regulators in the basal-like subtype, ARHGAP11A and RACGAP1 (Fig. 1A and B). Similarly to the mRNA levels of these genes in human tumors (Fig. 1C), protein expression of ArhGAP11A and RacGAP1 in human breast cancer cell lines was generally higher in those of the basal-like subtype than in other subtypes (Fig. 1D, Supplementary Fig. S2A). The basal-like cell lines SUM149 and HCC1937 both exhibited high expression of ArhGAP11A and RacGAP1 (Fig. 1D), and we therefore used these cell lines to determine the biological function(s) of these proteins in BLBC.
Figure 1.
ArhGAP11A and RacGAP1 are highly expressed in BLBC. A-B, the expression of Rho GEF, GAP, GTPase, and GDI genes in basal-like breast tumors is shown relative to their expression in (A) normal or (B) luminal A tumors, as determined from TCGA RNA-Seq data. The 24 genes with the highest relative expression in the basal-like subtype are shown. C, TCGA RNA-Seq data showing row median centered expression levels of ARHGAP11A and RACGAP1 mRNA in human breast tumors of different subtype. D, blot analyses for ArhGAP11A and RacGAP1 expression in human breast cancer cell lines of different subtypes. Actin expression was determined to verify equivalent loading of total protein.
ArhGAP11A and RacGAP1 are both required for BLBC cell line proliferation
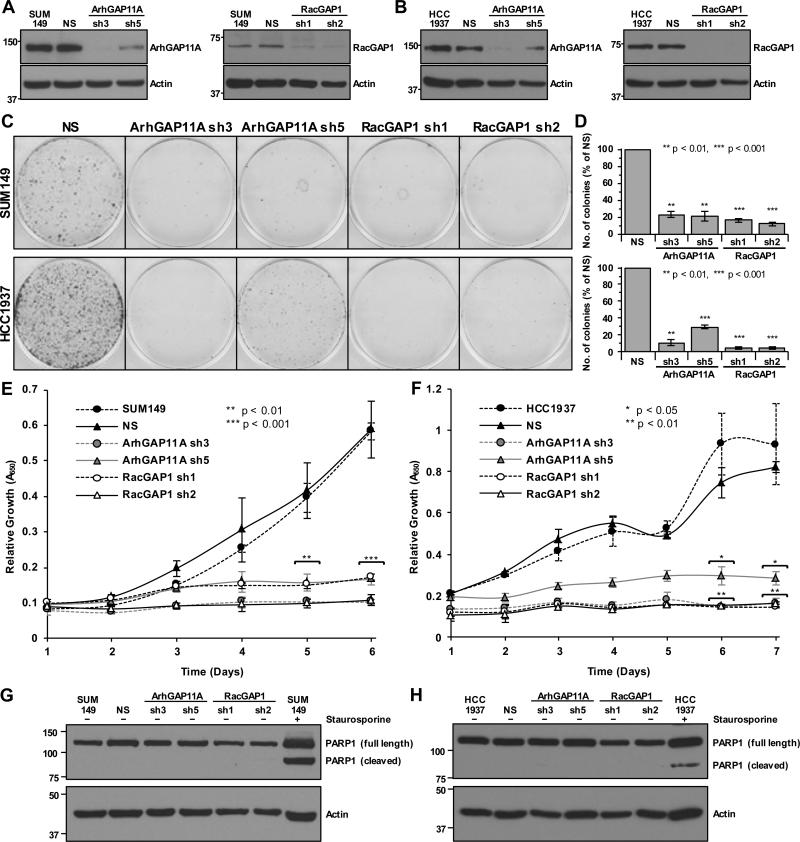
To assess the contribution of ArhGAP11A and RacGAP1 to BLBC cell growth, we stably suppressed their expression in SUM149 and HCC1937 cells using two separate lentivirus-delivered shRNA constructs per gene. In both cell lines, steady-state ArhGAP11A protein expression was consistently reduced by ~90% and ~60% by the sh3 and sh5 vectors, respectively, relative to NS or uninfected (labeled as SUM149 or HCC1937) control cells (Fig. 2A and B, Supplementary Fig. S1). For RacGAP1, sh1 and sh2 constructs both caused ~90% knockdown (Fig. 2A and B, Supplementary Fig. S1).
Figure 2.
ArhGAP11A and RacGAP1 are both required for BLBC proliferation. A-B, representative western blots showing knockdown of ArhGAP11A (left panels) and RacGAP1 (right panels) in (A) SUM149 and (B) HCC1937 cells. C, representative images of crystal violet-stained clonogenic growth assays with SUM149 (upper) and HCC1937 (lower) cells, with or without knockdown of ArhGAP11A or RacGAP1. D, quantification of mean colony formation for SUM149 (upper, n = three independent experiments performed in triplicate ±SEM) and HCC1937 (lower, n = three independent experiments performed in triplicate ±SEM) cells, with or without knockdown of ArhGAP11A or RacGAP1, normalized to NS control. Statistical significance was determined by one-sample t test relative to the NS control. E-F, MTT assays showing mean proliferation of (E) SUM149 and (F) HCC1937 cells, with or without knockdown of ArhGAP11A or RacGAP1, each from two independent experiments performed in triplicate ±SEM. Statistical significance, relative to the NS control, was determined by one-way ANOVA with Dunnett's post-hoc test. G-H, blot analyses showing PARP1 cleavage in (G) SUM149 and (H) HCC1937 cells, with or without knockdown of ArhGAP11A or RacGAP1, or after treatment with 1 μM staurosporine. Actin expression was determined to verify equivalent loading of total protein. Data shown are representative of three independent experiments.
We then performed a clonogenic growth assay to determine the effect of reduced ArhGAP11A or RacGAP1 expression on anchorage-dependent proliferation. SUM149 or HCC1937 cells depleted of ArhGAP11A or RacGAP1 formed ~70-95% fewer colonies of proliferating cells relative to the NS control (Fig. 2C and D). Near-complete suppression of proliferation was also observed for each RhoGAP using an MTT viability assay (Fig. 2E and F). The failure of BLBC cell lines to proliferate upon depletion of ArhGAP11A or RacGAP1 suggests that these GAPs act to promote cancer cell growth. Therefore, our results support the hypothesis that ArhGAP11A and RacGAP1 have oncogenic, rather than tumor suppressive, roles in BLBC cells. Depletion of these proteins from HER2-enriched (BT474) or luminal B (MCF7 and T47D) human breast cancer cell lines also caused defective proliferation (Supplementary Fig. S2), indicating that the importance of ArhGAP11A and RacGAP1 to breast cancer growth is not restricted to the BLBC subtype alone.
RacGAP1-knockdown results in cytokinesis failure
We next sought to delineate the mechanism(s) through which ArhGAP11A and RacGAP1 suppression impaired BLBC growth. Unlike in the presence of the apoptosis-inducing compound staurosporine, depletion of ArhGAP11A or RacGAP1 from SUM149 or HCC1937 cells did not result in detectable levels of cleaved PARP1, a marker for cell death (Fig. 2G and H). Hence, increased apoptosis was not responsible for the proliferation defects that arose upon knockdown of ArhGAP11A or RacGAP1.
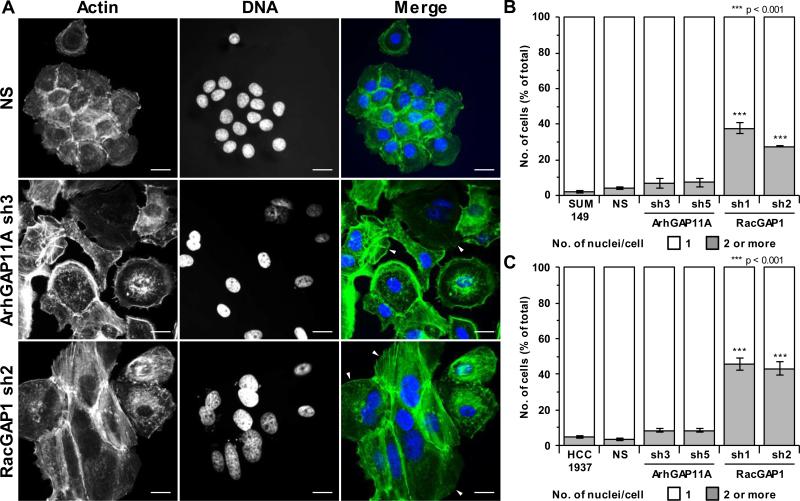
RacGAP1 has a well-documented role in regulating cytokinesis (29,30), and ArhGAP11A has also recently been implicated in the control of this process (31). We therefore examined the effect of GAP-knockdown on the efficiency of BLBC cell division. Fluorescence microscopy of Hoechst- and phalloidin-stained cells was used to identify bi- or multinucleated cells, which form upon cytokinesis failure. Consistent with the established role of RacGAP1 in regulating cytokinesis, ~30-40% of SUM149 cells, and ~45% of HCC1937 cells became bi/multinucleated upon RacGAP1-knockdown (Fig. 3A, B, and C). Hence, cytokinesis failure is likely to contribute to the inability of RacGAP1-depleted cells to proliferate. In contrast, knockdown of ArhGAP11A only resulted in ~7-9% of cells becoming bi/multinucleated (Fig. 3A, B, and C). Although this slight defect may partially contribute to growth impairment, it is insufficient to account for the substantial growth defects observed upon depletion of ArhGAP11A (Fig. 2C, D, E, and F).
Figure 3.
RacGAP1-knockdown results in cytokinesis failure. A, representative fluorescent images showing actin (green) and Hoechst (blue) staining of SUM149 cells with or without knockdown of ArhGAP11A or RacGAP1. Scale bar = 20 μm. Arrowheads in merged images indicate binucleated cells. B-C, quantification of the percentage of bi/multinucleated (B) SUM149 or (C) HCC1937 cells with or without knockdown of ArhGAP11A or RacGAP1 (n = three independent experiments in each case ±SEM, ≥ 108 total cells/condition/experiment). Statistical significance, relative to the NS control, was determined by one-way ANOVA with Dunnett's post-hoc test.
ArhGAP11A-depleted cells undergo p27-mediated cell cycle arrest
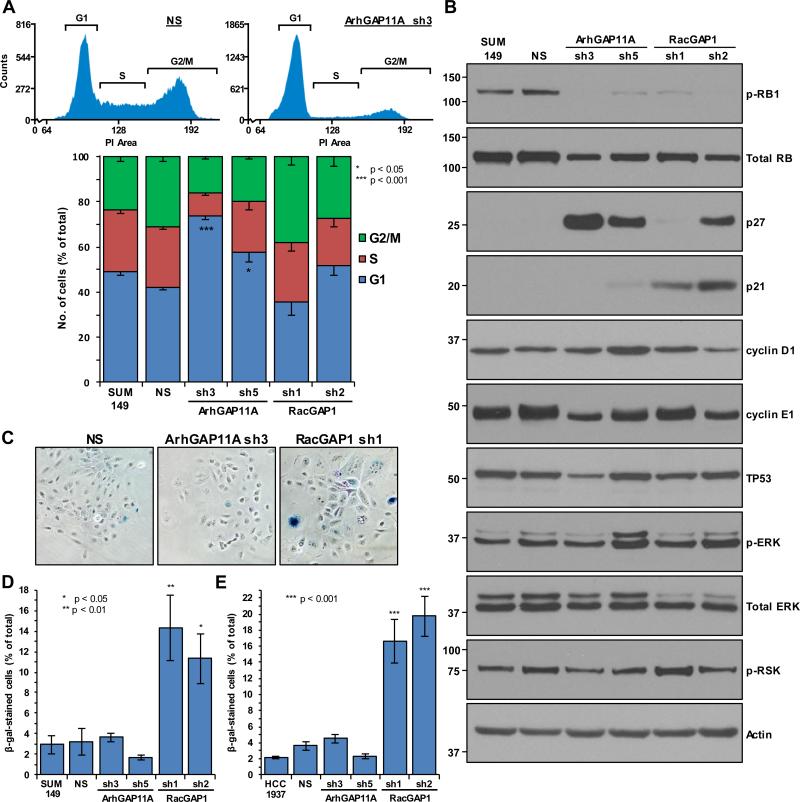
To gain further insight into the cause of proliferation failure in ArhGAP11A- and RacGAP1-depleted cells, we next performed flow cytometry analysis of propidium iodide-stained SUM149 cells to identify whether GAP-depletion caused defects in cell cycle progression. Relative to NS cells, ArhGAP11A-deficient cells accumulated in the G1 phase of the cell cycle (Fig. 4A). Hence, ArhGAP11A is required for efficient cell cycle progression in BLBC cells, indicative of an oncogenic role for this GAP.
Figure 4.
ArhGAP11A-depleted cells undergo p27-mediated cell cycle arrest whereas knockdown of RacGAP1 causes senescence. A, shown in the upper panels are representative histograms indicating the fluorescence intensity of propidium iodide (PI)-stained NS- or ArhGAP11A sh3-treated SUM149 cells, as identified using flow cytometry. The relative boundaries of the G1, S, and G2/M phases are indicated. Shown in the lower panel is quantification of the mean percentage of SUM149 cells in each phase of the cell cycle, with or without knockdown of ArhGAP11A or RacGAP1, from three independent experiments performed in triplicate ±SEM. Statistical significance for the G1 category is shown relative to the NS control, and was determined by one-way ANOVA with Dunnett's post-hoc test. B, blot analyses for the indicated proteins, from one experiment, representative of three. Actin expression was determined to verify equivalent loading of total protein. C, representative images of senescence-associated β-galactosidase (β-gal)-stained SUM149 cells (in blue), with or without knockdown of ArhGAP11A or RacGAP1. D-E, quantification of the mean percentage of β-gal-stained (D) SUM149 or (E) HCC1937 cells, with or without knockdown of ArhGAP11A or RacGAP1 (n = three independent experiments performed in triplicate in each case ±SEM, > 400 total cells/condition/experiment). Statistical significance, relative to the NS control, was determined by one-way ANOVA with Dunnett's post-hoc test.
To elucidate the molecular mechanisms responsible for causing G1 arrest in ArhGAP11A-depleted SUM149 cells, we determined the expression levels of proteins that regulate G1 to S phase cell cycle transition. Notably, phosphorylation and inactivation of the RB1 tumor suppressor protein, which promotes G1 to S progression by releasing the inhibition of E2F transcription factors, was dramatically reduced upon suppression of ArhGAP11A expression (Fig. 4B). RB1 phosphorylation is dependent on cyclin-dependent kinases (CDKs) in complex with cyclins D1 and E, but can be prevented by CDK inhibitors such as CDKN2A/p16, p21, or p27. After knockdown of ArhGAP11A, strong induction of expression of p27 was detected in SUM149 cells (Fig. 4B). As neither p21 (Fig. 4B) nor p16 (which is deleted in SUM149 cells (32)) were detected under the same conditions, these results indicate that p27 is the CDK inhibitor responsible for the hypophosphorylation of RB1 and the associated G1 arrest that occurs upon depletion of ArhGAP11A. No substantial changes were observed in the expression levels of any of the other cell cycle regulators that we tested (Fig. 4B). We suggest that p27-mediated arrest is likely to be the major mechanism through which ArhGAP11A-depleted cells fail to proliferate.
RacGAP1-knockdown causes senescence
In contrast to ArhGAP11A, knockdown of RacGAP1 from SUM149 cells did not alter the cell cycle profile relative to control cells (Fig. 4A). Despite this, RacGAP1-depleted cells also exhibited low levels of RB1 phosphorylation (Fig. 4B). In these cells, the CDK inhibitor p21 was upregulated (Fig. 4B), indicating that different pathways are activated in response to the depletion of RacGAP1 as compared to ArhGAP11A. As p21-mediated inhibition of RB1 phosphorylation is a pathway known to induce senescence (33), we next tested to see whether senescence may contribute to the growth defect of RacGAP1-deficient cells. Indeed, SUM149 and HCC1937 cells depleted of RacGAP1 were found to have enhanced levels of senescence-associated β-galactosidase expression (Fig. 4C, D, and E). In contrast, ArhGAP11A-knockdown did not induce senescence (Fig. 4C, D, and E), providing further support for the distinct roles that ArhGAP11A and RacGAP1 play in BLBC cells: depletion of ArhGAP11A leads to decreased growth via p27-mediated cell cycle arrest, whereas RacGAP1-deficient cells fail to proliferate as a result of the combined effects of cytokinesis failure, p21-induction, and the onset of senescence.
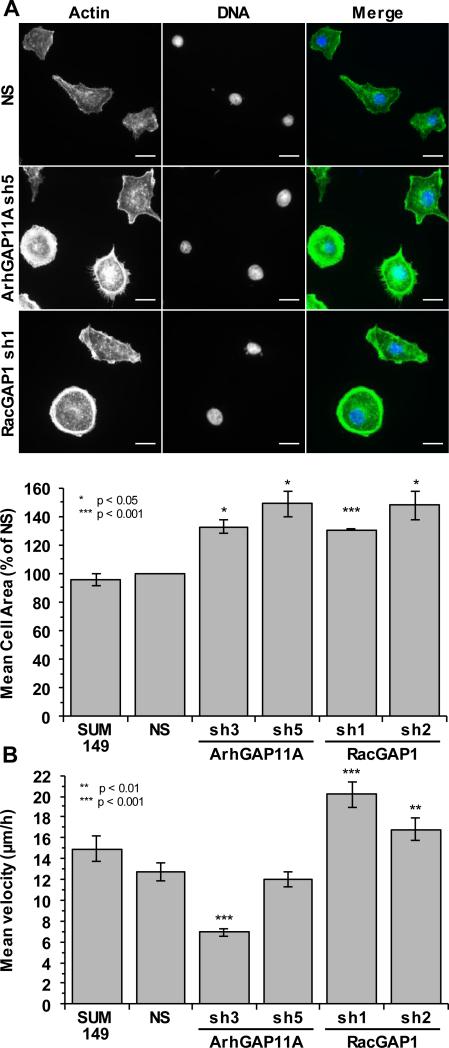
ArhGAP11A and RacGAP1 regulate cell spreading and migration
In addition to determining the consequences of depletion of ArhGAP11A and RacGAP1 on BLBC proliferation, we also examined their roles in regulating cell spreading and migration, two processes known to be reliant on Rho GTPase-dependent cytoskeletal dynamics and important for invasive and metastatic cancer growth (34). Suppression of the expression of either ArhGAP11A or RacGAP1 caused mononucleated SUM149 cells to spread on fibronectin with an approximately 30-50% larger area compared to that of NS control cells (Fig. 5A). Enhanced spreading was also observed on uncoated glass coverslips (Supplementary Fig. S3). As bi/multinucleated cells typically exhibit greatly increased spread areas, these cells were excluded from the spreading analysis. These results indicate that both ArhGAP11A and RacGAP1 are involved of the control of cell spreading, suggesting that one function of these GAPs may be to regulate the cytoskeleton via Rho GTPase signaling. However, the possibility that the spread morphologies of these cells may be secondarily linked to the onset of senescence, particularly in the case of RacGAP1-depleted cells, cannot be excluded.
Figure 5.
ArhGAP11A and RacGAP1 regulate cell spreading and migration. A, shown in the upper panel are representative fluorescent images of actin (green) and Hoechst (blue) staining of SUM149 cells, with or without knockdown of ArhGAP11A or RacGAP1, after 2 h on 10 μg/ml fibronectin-coated coverslips. Scale bar = 20 μm. Shown in the lower panel is quantification of mean cell area (n = three independent experiments ±SEM, ≥85 cells/condition/experiment) after spreading on fibronectin, normalized to NS control. Statistical significance was determined by one-sample t test relative to the NS control. Multinucleated cells were excluded from the analysis. B, mean random migration velocity of SUM149 cells, with or without knockdown of ArhGAP11A or RacGAP1, on fibronectin-coated coverslips, as measured from 24 h time-lapse movies. Data shown were pooled from three independent experiments, ≥110 total cell tracks/condition ±SEM. Statistical significance, relative to the NS control, was determined by one-way ANOVA with Dunnett's post-hoc test.
The effect of ArhGAP11A and RacGAP1 on random migration was assessed using time-lapse microscopy of cells plated on fibronectin. Tracking the movement of individual cells over a 24 h period determined that SUM149 cells treated with ArhGAP11A sh3 had a significantly (~45%) reduced mean velocity compared to that of NS cells (Fig. 5B). This migratory defect was not observed upon treatment with the ArhGAP11A sh5 construct, most likely reflecting the enhanced efficiency of ArhGAP11A knockdown with sh3 compared to sh5 (Supplementary Fig. S1I). Surprisingly, the mean migration velocity of RacGAP1-depleted cells was 32-58% faster than that of NS cells (Fig. 5B). These results indicate that ArhGAP11A promotes, whereas RacGAP1 inhibits, BLBC cell migration in vitro.
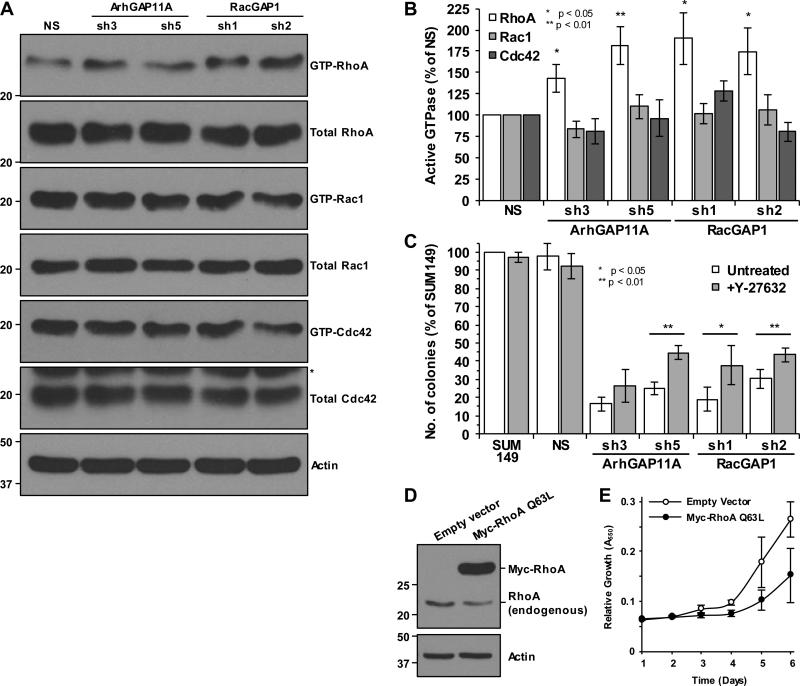
RhoA activity is increased upon depletion of ArhGAP11A or RacGAP1
As proliferation, cytokinesis, cell cycle progression, spreading, and migration – processes that we have shown to be affected by ArhGAP11A and/or RacGAP1 in BLBC cells – are known to be directly regulated by Rho GTPases, we next set out to determine which specific Rho GTPases are controlled by these two GAPs. In SUM149 cells, pulldown experiments for active GTPases revealed that RhoA, but not Rac1 or Cdc42, was more active upon depletion of ArhGAP11A than in NS-treated cells, by an average of 43-82% (Fig. 6A and B). This is consistent with ArhGAP11A having catalytic GAP activity toward RhoA, as has been demonstrated using in vitro assays (31,35,36). In contrast, RacGAP1 has previously been demonstrated to act as a GAP for Rac1 and Cdc42, but not RhoA in vitro (37). To our surprise, Rho GTPase pulldown experiments showed that RhoA activity, but not that of Rac1 or Cdc42, was elevated in SUM149 cells upon depletion of RacGAP1 (Fig. 6A and B). These results suggest that RhoA activity in BLBC cells is usually suppressed by ArhGAP11A or RacGAP1. To assess whether increased RhoA activity is responsible for the proliferation defect of ArhGAP11A- or RacGAP1-depleted cells, we performed clonogenic growth assays in the presence of the ROCK protein kinase-selective inhibitor Y-27632. ROCK is a downstream effector of RhoA and its inhibition caused a partial rescue of the growth defect of ArhGAP11A- or RacGAP1-depleted cells (Fig. 6C), indicating that efficient proliferation is dependent on RhoA signaling being restricted. Furthermore, transfection of SUM149 cells with a constitutively active RhoA mutant (Q63L) resulted in decreased proliferation (Fig. 6D and E), consistent with observations made with another constitutively active RhoA mutant (G14V) in Swiss3T3 fibroblasts (38). For comparison, constitutively active Rac1 and Cdc42 mutants (Q61L) had no effect on proliferation (Supplementary Fig. S4). These results emphasize that persistently elevated RhoA activity is prohibitive to growth.
Figure 6.
RhoA activity is increased upon depletion of ArhGAP11A or RacGAP1. A, blot analyses for GTP-bound RhoA, Rac1, and Cdc42 levels in SUM149 cells, with or without knockdown of ArhGAP11A or RacGAP1, following Rho GTPase pulldown experiments. Total protein levels were detected from whole cell lysate (*non-specific band from previous blot). Actin expression was determined to verify equivalent loading of total protein. B, densitometric quantification of the mean ratio of GTP-bound to total RhoA, Rac1, or Cdc42 (±SEM), from seven, eight, or four independent pulldown experiments, respectively. Data were normalized to the NS control in each case. Statistical significance was determined by one-sample t test relative to the NS control. C, quantification of mean SUM149 colony formation, with or without knockdown of ArhGAP11A or RacGAP1, and after treatment with or without 10 μM Y-27632 (n = four independent experiments performed in triplicate ±SEM, normalized to untreated SUM149 cells). Statistical significance between untreated and Y-27632-treated samples was determined by two-tailed t test. D, blot analyses for RhoA showing endogenous RhoA and overexpression of Myc-RhoA Q63L in SUM149 cells. Actin expression was determined to verify equivalent loading of total protein. E, MTT assay showing SUM149 cell proliferation in the presence or absence of Myc-RhoA Q63L (n = two independent experiments performed in triplicate ±SEM).
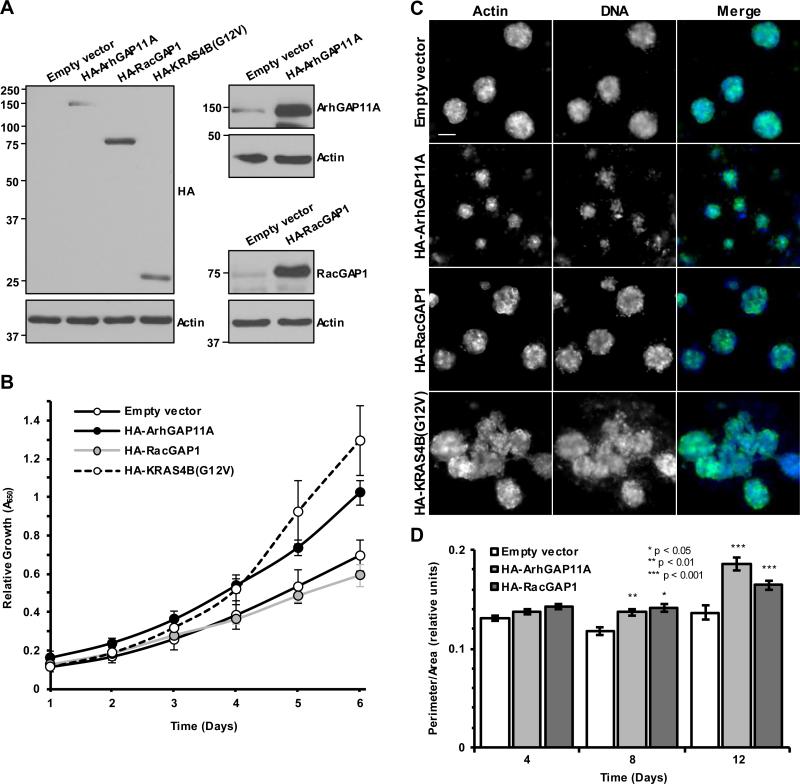
ArhGAP11A or RacGAP1 expression elicits oncogenic phenotypes in untransformed cells
To assess the effect of ArhGAP11A and RacGAP1 on transformation, we stably overexpressed these proteins in untransformed immortalized human MCF10A breast myoepithelial cells (Fig. 7A). In MTT assays, expression of ArhGAP11A resulted in enhanced MCF10A proliferation, as did expression of a known oncogene, mutationally-activated KRAS4B(G12V) (Fig. 7B). These data support the idea that ArhGAP11A can act as an oncogene and cancer driver. Interestingly, overexpression of HA-RacGAP1 did not cause an increase in proliferation relative to empty vector control (Fig. 7B). It is possible that other signaling components must also be altered for RacGAP1 to enhance proliferation.
Figure 7.
ArhGAP11A or RacGAP1 expression elicits oncogenic phenotypes in untransformed cells. A, blot analyses for HA-epitope tag (left panels), ArhGAP11A (upper right panels), and RacGAP1 (lower right panels) showing overexpression of HA-ArhGAP11A, HA-RacGAP1, and HAKRAS4B(G12V) in MCF10A cells. Actin expression was determined to verify equivalent loading of total protein. B, MTT assay to monitor MCF10A proliferation in empty vector control cells or cells expressing HA-ArhGAP11A, HA-RacGAP1, or HA-KRAS4B(G12V) (n = three independent experiments performed in triplicate ±SEM). C, representative images of actin (green) and Hoechst (blue) stained MCF10A acini expressing empty vector control, HA-ArhGAP11A, HA-RacGAP1, or HA-KRAS4B(G12V), after 12 days growth on Matrigel. Scale bar = 50 μm. D, quantification of relative acini perimeter/area over time for MCF10A cells expressing empty vector, HA-ArhGAP11A, or RacGAP1. Data shown were pooled from two independent experiments performed in duplicate, ≥161 total acini/day/condition ±SEM. Statistical significance, relative to the empty vector control, was determined by one-way ANOVA with Dunnett's post-hoc test.
We next performed MCF10A acinar formation assays to evaluate the effect of RhoGAP overexpression on mammary epithelial morphogenesis. The expression of oncogenes in MCF10A cells can lead to the formation of disrupted acinar morphology; for example, KRAS4B(G12V) expression resulted in the formation of large multi-acinar structures (Fig. 7C). Acini ectopically overexpressing either ArhGAP11A or RacGAP1 exhibited a disrupted, less spherical architecture relative to control acini as early as 8 days after plating (Fig. 7C and D). This is consistent with an oncogenic effect for both ArhGAP11A and RacGAP1.
Discussion
Our RNA-Seq analysis of human breast tumors identified high expression of several RhoGAP genes in the basal-like subtype, raising the possibility that these negative regulators of Rho GTPases can act as oncoproteins rather than tumor suppressors in BLBC. To address this, we focused on two RhoGAPs that are highly expressed in BLBC, ArhGAP11A and RacGAP1, and established that they are essential for the proliferation of basal-like cell lines. The mechanisms through which these GAPs promote growth differ, in that ArhGAP11A is required for cell cycle progression, whereas cytokinesis is dependent on RacGAP1. Despite the divergent functions of these GAPs, both inhibit RhoA. Hence, our results not only suggest that RhoGAPs have an oncogenic role in BLBC, but also support recent observations that RhoA may act as a tumor suppressor.
While certain RhoGAPs, most notably DLC1 (14,15), have been characterized and well-validated as tumor suppressors, our findings indicate that this classification is not applicable to all members of the RhoGAP family, as had previously been assumed. Compared to RhoGEFs, RhoGAPs are relatively poorly characterized, particularly with regard to their role in cancer. However, examples of other RhoGAPs having pro-tumorigenic functions in breast cancer do exist, such as ArhGAP35 (39), ArhGAP5 (40,41), and ArhGAP31 (42). These studies, coupled to our own, provide accumulating evidence that the role of the RhoGAP family in cancer is not as straightforward as might be expected from the classical interpretation of Rho GTPases as oncogenes. However, our results appear less paradoxical in light of recent studies that have identified RhoA mutations in cancer that are consistent with reduced activity of this GTPase (16-21). If reduced RhoA activity is indeed advantageous for cancer cell proliferation, then it follows that regulation by GAPs would be a means of achieving this. Given the diverse functions of RhoA, it is possible that RhoGAPs such as ArhGAP11A and RacGAP1 permit RhoA activity to be decreased in a very precise spatiotemporal fashion, such that RhoA can still perform other roles within the cell. This could potentially explain why missense mutations and deletions of RhoA are relatively rare in breast cancer (<1.5%) (28).
Our results indicate that ArhGAP11A is required for G1 to S phase cell cycle progression in BLBC cells. Indeed, depletion of ArhGAP11A caused G1 arrest associated with increased p27 expression. ArhGAP11A has previously been implicated in the control of cytokinesis, with 18% of HeLa cells failing this process upon its knockdown (31). In SUM149 and HCC1937 BLBC cell lines, we did not observe strong defects in cytokinesis upon depletion of ArhGAP11A, however. This discrepancy may reflect heterogeneity between cancer cell lines, and suggests that ArhGAP11A may have additional, non-cytokinesis-related functions in BLBC. In HCT116 colon cancer cells, ArhGAP11A has been shown to regulate invasion, but knockdown of this GAP was not reported to affect cell proliferation (35). Despite this, depletion of ArhGAP11A reduced tumor growth in vivo (35), which would appear to agree with our finding that cells deficient of this GAP had substantial defects in proliferation. High expression of ArhGAP11A has also been reported in colorectal, brain, and lung cancers (35), suggesting that its pro-tumorigenic role may not just be limited to breast cancer. Furthermore, a truncated paralog of ArhGAP11A called ArhGAP11B (which is also highly expressed in BLBC, Fig. 1A and B), has recently been shown to promote neocortex expansion (43), raising the intriguing possibility that pro-proliferative functions may be conserved between these related proteins.
Elevated RacGAP1 expression has also been reported in a variety of cancers, including breast, and its overexpression is frequently correlated to poor patient survival and recurrence (44-49). Its biological function in cancer is poorly understood, however. In BLBC, we found that RacGAP1-depleted cells failed to proliferate as a result of cytokinesis failure, p21-induction, and the onset of senescence. The role of RacGAP1 in regulating cytokinesis is well-established (29,30) and, in BLBC, high levels of this protein may be required to facilitate excessive cell division. The GTPase specificity of RacGAP1 is controversial, with one study suggesting that this GAP could be converted from a Rac1- to a RhoA-specific GAP upon phosphorylation by aurora kinase B (50). Others have refuted this finding, however (37). In BLBC cells, similarly to in squamous cell carcinoma cells (46), knockdown of RacGAP1 caused an increase in cellular RhoA activity. It is possible that RhoA activity may be indirectly altered by the depletion of RacGAP1, for example as a result of mislocalized Ect2 – a RhoA-selective RhoGEF whose localization is otherwise regulated by RacGAP1 (30).
Despite depletion of either ArhGAP11A or RacGAP1 leading to increased RhoA activity, we observed distinct mechanisms of growth suppression in each case. These differences may be caused by GAP catalytic-independent functions, discrete spatiotemporal regulation of RhoA signaling, or through additional GAP activity on other Rho GTPase substrates. The only known catalytic domain found in ArhGAP11A and RacGAP1 is a RhoGAP domain but, through their divergent flanking sequences, they are likely to possess scaffolding functions and interact with distinct sets of proteins. RacGAP1 is a component of the centralspindlin complex during mitosis and cytokinesis, but its function during interphase is not well established. Similarly, little is known about the subcellular localization of ArhGAP11A. In future studies, it would be of interest to characterize the localization of ArhGAP11A and RacGAP1 in BLBC cells to identify whether this influences their distinct abilities to act as oncogenes. Furthermore, a comprehensive biochemical analysis of their GTPase substrates, beyond the three that are conventionally evaluated (RhoA, Rac1, and Cdc42), would provide additional insight into their GAP-dependent mechanisms of action.
Supplementary Material
Acknowledgments
CDL was supported by the U.S. Army Medical Research and Materiel Command under Award No. W81XWH-14-1-0033. Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the U.S. Army. This work was also supported by National Institutes of Health grants to CJD (CA042978, CA179193, and CA175747), to KB (GM029860 and GM103723), and to CMP (NCI Breast SPORE program P50-CA58223-09A1). We thank Tikvah Hayes for providing the pCDH HA-KRAS4B(G12V) construct and Janet Dow for help with flow cytometry. The UNC Flow Cytometry Core Facility is supported in part by P30-CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Financial Support: C.J. Der was also supported by a Pancreatic Cancer Action Network-AACR RAN Grant.
Footnotes
Conflicts of interest: C.M. Perou is on the board of directors, has ownership interest (including patents), and is a consultant/advisory board member for bioclassifier LLC. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–33. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 6.Crown J, O'Shaughnessy J, Gullo G. Emerging targeted therapies in triple-negative breast cancer. Ann Oncol. 2012;23(Suppl 6):vi56–65. doi: 10.1093/annonc/mds196. [DOI] [PubMed] [Google Scholar]
- 7.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–6. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 8.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–12. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orgaz JL, Herraiz C, Sanz-Moreno V. Rho GTPases modulate malignant transformation of tumor cells. Small GTPases. 2014;5:e29019. doi: 10.4161/sgtp.29019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay CR, Lawn S, Campbell AD, Faller WJ, Rambow F, Mort RL, et al. P-Rex1 is required for efficient melanoblast migration and melanoma metastasis. Nat Comm. 2011;2:555. doi: 10.1038/ncomms1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Csepanyi-Komi R, Safar D, Grosz V, Tarjan ZL, Ligeti E. In silico tissue-distribution of human Rho family GTPase activating proteins. Small GTPases. 2013;4:90–101. doi: 10.4161/sgtp.23708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–57. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukasik D, Wilczek E, Wasiutynski A, Gornicka B. Deleted in liver cancer protein family in human malignancies (Review). Oncol Lett. 2011;2:763–68. doi: 10.3892/ol.2011.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popescu NC, Goodison S. Deleted in liver cancer-1 (DLC1): an emerging metastasis suppressor gene. Mol Diagn Ther. 2014;18:293–302. doi: 10.1007/s40291-014-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–70. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–5. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 18.Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:371–5. doi: 10.1038/ng.2916. [DOI] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–7. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 21.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 22.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahai E, Alberts AS, Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 1998;17:1350–61. doi: 10.1093/emboj/17.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues P, Macaya I, Bazzocco S, Mazzolini R, Andretta E, Dopeso H, et al. RHOA inactivation enhances Wnt signalling and promotes colorectal cancer. Nat Comm. 2014;5:5458. doi: 10.1038/ncomms6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wennerberg K, Ellerbroek SM, Liu RY, Karnoub AE, Burridge K, Der CJ. RhoG signals in parallel with Rac1 and Cdc42. J Biol Chem. 2002;277:47810–7. doi: 10.1074/jbc.M203816200. [DOI] [PubMed] [Google Scholar]
- 26.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 27.Wittchen ES, Burridge K. Analysis of low molecular weight GTPase activity in endothelial cell cultures. Methods Enzymol. 2008;443:285–98. doi: 10.1016/S0076-6879(08)02014-4. [DOI] [PubMed] [Google Scholar]
- 28.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–19. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo Y, Oh W, Frost JA. Controlling the switches: Rho GTPase regulation during animal cell mitosis. Cell Signal. 2014;26:2998–3006. doi: 10.1016/j.cellsig.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao WM, Fang G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc Natl Acad Sci U S A. 2005;102:13158–63. doi: 10.1073/pnas.0504145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanin E, Desai A, Poser I, Toyoda Y, Andree C, Moebius C, et al. A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev Cell. 2013;26:496–510. doi: 10.1016/j.devcel.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollestelle A, Nagel JH, Smid M, Lam S, Elstrodt F, Wasielewski M, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat. 2010;121:53–64. doi: 10.1007/s10549-009-0460-8. [DOI] [PubMed] [Google Scholar]
- 33.Campisi J. Aging, cellular senescence, and cancer. Ann Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawson CD, Burridge K. The on-off relationship of Rho and Rac during integrin-mediated adhesion and cell migration. Small GTPases. 2014;5:e27958. doi: 10.4161/sgtp.27958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagawa Y, Matsumoto S, Kamioka Y, Mimori K, Naito Y, Ishii T, et al. Cell cycle-dependent Rho GTPase activity dynamically regulates cancer cell motility and invasion in vivo. PLoS One. 2013;8:e83629. doi: 10.1371/journal.pone.0083629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Zhou X, Wang J, Li Z, Kong X, Qian J, et al. RhoGAPs attenuate cell proliferation by direct interaction with p53 tetramerization domain. Cell Rep. 2013;3:1526–38. doi: 10.1016/j.celrep.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 37.Bastos RN, Penate X, Bates M, Hammond D, Barr FA. CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis. J Cell Biol. 2012;198:865–80. doi: 10.1083/jcb.201204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin P, Flors C, Olson MF. Constitutively active RhoA inhibits proliferation by retarding G(1) to S phase cell cycle progression and impairing cytokinesis. Eur J Cell Biol. 2009;88:495–507. doi: 10.1016/j.ejcb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen CH, Chen HY, Lin MS, Li FY, Chang CC, Kuo ML, et al. Breast tumor kinase phosphorylates p190RhoGAP to regulate rho and ras and promote breast carcinoma growth, migration, and invasion. Cancer Res. 2008;68:7779–87. doi: 10.1158/0008-5472.CAN-08-0997. [DOI] [PubMed] [Google Scholar]
- 40.Heckman-Stoddard BM, Vargo-Gogola T, McHenry PR, Jiang V, Herrick MP, Hilsenbeck SG, et al. Haploinsufficiency for p190B RhoGAP inhibits MMTV-Neu tumor progression. Breast Cancer Res. 2009;11:R61. doi: 10.1186/bcr2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McHenry PR, Sears JC, Herrick MP, Chang P, Heckman-Stoddard BM, Rybarczyk M, et al. P190B RhoGAP has pro-tumorigenic functions during MMTV-Neu mammary tumorigenesis and metastasis. Breast Cancer Res. 2010;12:R73. doi: 10.1186/bcr2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Y, Northey JJ, Primeau M, Machado RD, Trembath R, Siegel PM, et al. CdGAP is required for transforming growth factor beta- and Neu/ErbB-2-induced breast cancer cell motility and invasion. Oncogene. 2011;30:1032–45. doi: 10.1038/onc.2010.477. [DOI] [PubMed] [Google Scholar]
- 43.Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–70. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 44.Pliarchopoulou K, Kalogeras KT, Kronenwett R, Wirtz RM, Eleftheraki AG, Batistatou A, et al. Prognostic significance of RACGAP1 mRNA expression in high-risk early breast cancer: a study in primary tumors of breast cancer patients participating in a randomized Hellenic Cooperative Oncology Group trial. Cancer Chemother Pharmacol. 2013;71:245–55. doi: 10.1007/s00280-012-2002-z. [DOI] [PubMed] [Google Scholar]
- 45.Imaoka H, Toiyama Y, Saigusa S, Kawamura M, Kawamoto A, Okugawa Y, et al. RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis. 2015;36:346–54. doi: 10.1093/carcin/bgu327. [DOI] [PubMed] [Google Scholar]
- 46.Hazar-Rethinam M, de Long LM, Gannon OM, Boros S, Vargas AC, Dzienis M, et al. RacGAP1 Is a Novel Downstream Effector of E2F7-Dependent Resistance to Doxorubicin and Is Prognostic for Overall Survival in Squamous Cell Carcinoma. Molecular cancer therapeutics. 2015;14:1939–50. doi: 10.1158/1535-7163.MCT-15-0076. [DOI] [PubMed] [Google Scholar]
- 47.Saigusa S, Tanaka K, Mohri Y, Ohi M, Shimura T, Kitajima T, et al. Clinical significance of RacGAP1 expression at the invasive front of gastric cancer. Gastric cancer : official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2015;18:84–92. doi: 10.1007/s10120-014-0355-1. [DOI] [PubMed] [Google Scholar]
- 48.Wang SM, Ooi LL, Hui KM. Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin Cancer Res. 2011;17:6040–51. doi: 10.1158/1078-0432.CCR-11-0557. [DOI] [PubMed] [Google Scholar]
- 49.Ma XJ, Salunga R, Dahiya S, Wang W, Carney E, Durbecq V, et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14:2601–8. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 50.Minoshima Y, Kawashima T, Hirose K, Tonozuka Y, Kawajiri A, Bao YC, et al. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell. 2003;4:549–60. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.