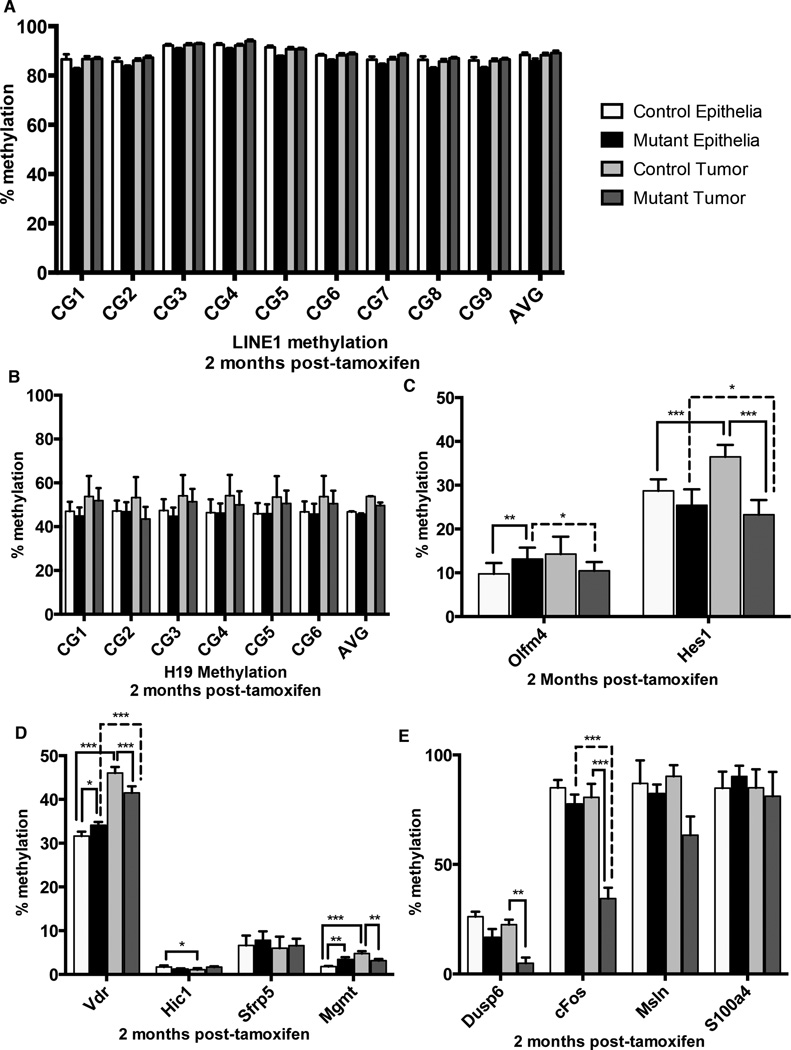
Figure 4. Dnmt1 mutant tumors show increased hypomethylation at potential oncogenes.
Targeted bisulfite sequencing was performed to estimate maintenance methylation of Dnmt1-deficient tumors, relative to controls tumors and to adjacent non-neoplastic crypt epithelium. Tumors and crypt epithelium were isolated by laser capture microdissection from controls and mutants two months following tamoxifen treatment.
(A–B) Analysis of nine CpGs in the LINE1 repetitive elements (NCBI Accession #D84391: 976-1,072) revealed no alterations in methylation of the repetitive transposable elements (A). Analysis of six CpGs in the H19 imprinting control region (Chr7: 149,766,621-149,766,690) showed that H19 methylation levels are comparable between Dnmt1 mutants and controls, in both tumor and non-tumor epithelium.
(C) Potential enhancer regions of genes involved in intestinal stem cell identity and proliferation, Olfm4 and Hes1 were analyzed for methylation changes in all conditions. Regional averages of seven CpGs in the Olfm4 enhancer (chr14:80399983-80400271) and six CpGs in the Hes1 enhancer (chr16:30055572-30055738) are shown. The Hes1 enhancer shows increased methylation in control tumors compared to adjacent tissue, which is abolished by Dnmt1 deletion.
(D) Genes with cancer-specific promoter hypermethylation, Vdr, Hic1, Sfrp5, Mgmt, were analyzed for methylation changes in all conditions. Regional averages of eight CpGs in the Vdr promoter (chr15:97731208-97731590), sixteen CpGs in Hic1 promoter (chr11:74983743-74983918), sixteen CpGs in the Sfrp5 promoter (chr19:42276328-42276453), and nineteen CpGs in Mgmt promoter (chr7:144086076-144086345) are shown. Only Vdr and Mgmt displayed significant hypermethylation in control tumors compared to adjacent tissue. Loss of Dnmt1 caused significant loss of hypermethylation at both loci suggesting hypermethylation of these genes is not required for tumor progression.
(E) Genes with cancer-specific promoter hypomethylation, Dusp6, c-Fos, Msln, S100a4, were analyzed for methylation changes in all conditions. Regional averages of eight CpGs in the Dusp6 promoter (chr10:98728738-98729028), fourteen CpGs in the c-Fos promoter (chr12:86817243-86817533), six CpGs in the Msln promoter (chr17:25891502-25891643), and three CpGs in the S100a4 promoter (chr3:90407319-90407489) are shown. Only Dusp6 and c-Fos displayed significant hypomethyation in control tumors compared to adjacent tissue. Dnmt1 deletion caused even more drastic loss of methylation at both promoters suggesting that loss of Dnmt1 may be driving activation of potential oncogenes during tumor growth.