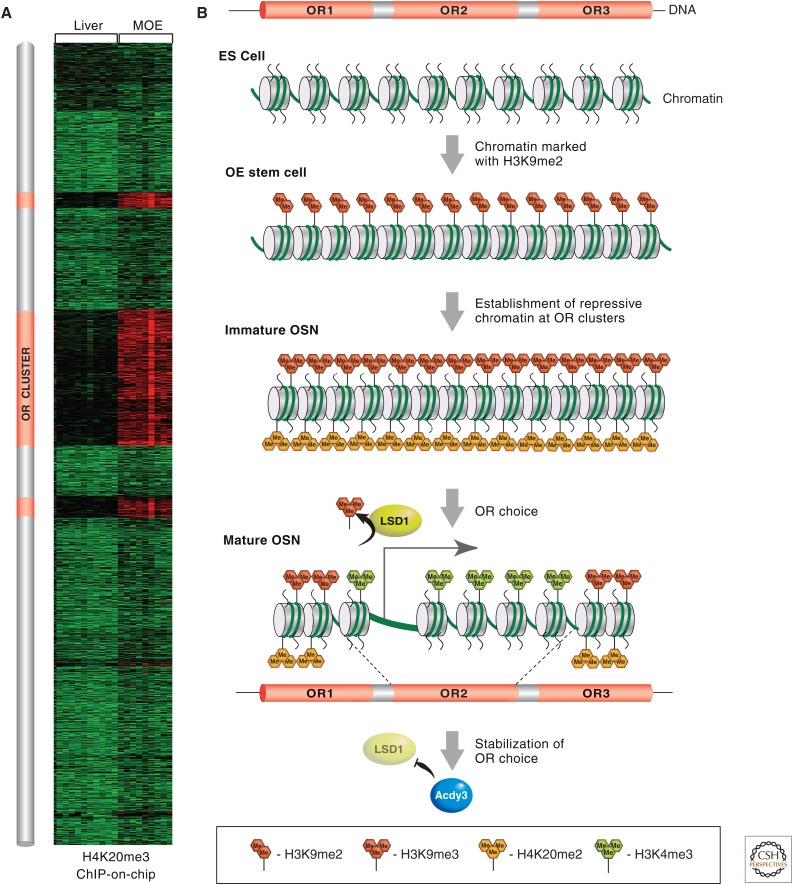
Figure 8.
MOE-specific epigenetic regulation of OR gene choice during OSN development. (A) A heatmap depicting various enrichment levels for H4K20me3 on all the genes of mouse chromosome 2, from ChIP-on-chip analysis from MOE or liver. Red reflects high enrichment levels and green shows no enrichment. The three “red clusters” of high H4K20me3 levels coincide with the genomic coordinates of the OR clusters, schematically represented on this chromosome in orange to the left of the heatmap. (B) OR clusters start in early development (ES cells), devoid of most histone marks. At the onset of olfactory neurogenesis, OR clusters are marked with H3K9me2. Most OR genes then become marked by H3K9me3 and H4K20me3 during the transition from basal pluripotent cells to OSN. The OR allele that is chosen for expression (i.e., OR2) becomes liberated from its repressive chromatin marks and remodeled into an active chromatin configuration, aided by the action of the H3K9 demethylase, LSD1, and becomes positive for H3K4me3. Toward the end of the OSN differentiation pathway, the locking in of a single active OR gene is facilitated by the OR protein-induced expression of adenylyl cyclase III (Adcy3), which inhibits further LSD1 histone demethylation.