FIGURE 2.
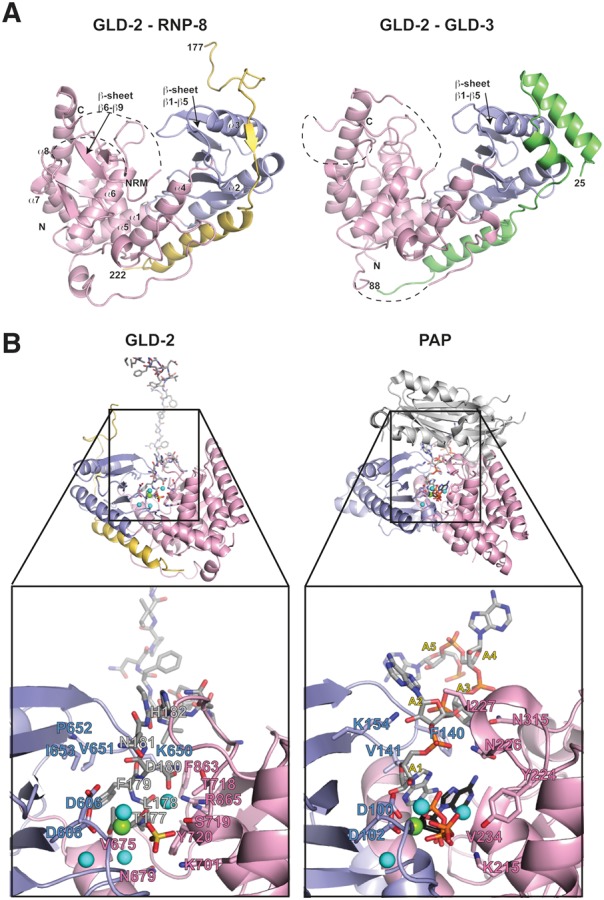
Structure of a GLD-2–RNP-8 core complex. (A) Ribbon diagram of the GLD-2PAP-D–RNP-8GB complex (PDB code 5JNB, left panel) and GLD-2PAP-D–GLD-3NT (PDB code 4ZRL, right panel) shown in the same orientation after optimal superposition of their central domains (in pink). The catalytic domains are in blue, RNP-8 in yellow, and GLD-3 in green. The N- and C-terminal residues of the proteins are labeled. Disordered loops are indicated with dashed lines. The five-stranded β-sheet (β1–β5) of the catalytic domain is indicated in both structures. In GLD-2PAP-D–RNP-8GB, an additional four-stranded β-sheet (β6–β9) is well ordered on top of the central domain. These secondary structure elements correspond to a conserved polypeptide segment between helices α7 and α8 and lay on top of helix α6. NRM, nucleotide recognition motif. (B) On the left is a zoom-in of the active site cleft of the GLD-2PAP-D–RNP-8GB complex shown after a 180° rotation around a vertical axis with respect to the view in panel A. The colors are the same as in panel A, with the peptide of a symmetry-related RNP-8 molecule (for explanations see main text) in stick representation and with carbon atoms in gray. On the right is the corresponding zoom-in view from the structure of canonical PAP bound to RNA and ATP (in stick representation, with carbon atoms in gray and black, respectively) (PDB code 2Q66, Balbo and Bohm 2007). For clarity, the zoom-in view lacks the RRM domain of canonical PAP (shown as a reference in the overall view in light gray). Magnesium and water molecules are shown as green and cyan spheres, respectively. A set of important residues in both GLD-2–RNP-8GB and PAP are highlighted in stick representation and labeled. Note that the place of the two most 3′ end ribonucleotides is taken by RNP-8 Phe179 and His182 (instead of the nucleotide bases) and Asn181 (instead of the nucleotide ribose moieties). The place of ATP is taken by RNP-8 Thr177, Leu178, and Asp180 (instead of the adenosine) and by Asp180 and a sulfate ion from the crystallization buffer (instead of the phosphates). Remarkably, the GLD-2PAP-D–RNP-8GB structure also shows a similar arrangement of magnesium and water molecules in the active site as observed in the PAP-RNA-ATP structure (Balbo and Bohm 2007).
