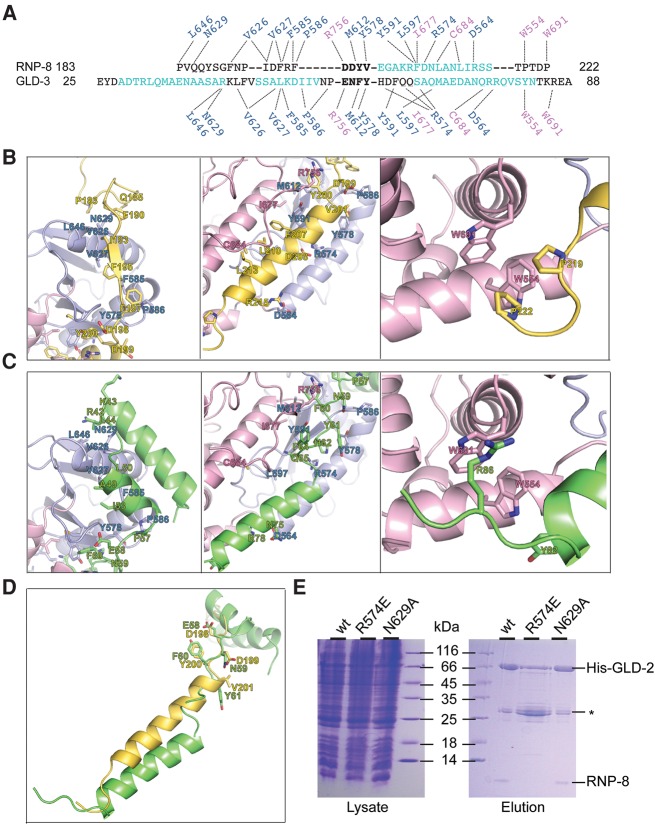
FIGURE 3.
Structural basis of mutually exclusive GLD-2–RNP-8 and GLD-2–GLD-3 complex formation. (A) Structural alignment from C. elegans RNP-8GB and GLD-3NT, with α-helical residues in cyan. Note that only the DDYV segment of RNP-8 and the ENFY segment of GLD-3 (both shown in bold letters) superpose well, with a C-α root mean square deviation of <0.1 Å. The dotted lines indicate the interactions of different GLD-3 and RNP-8 residues with the same side chains in the GLD-2 catalytic and central domains (in blue and pink above and below the sequences). (B) Zoom-in of RNP-8GB interactions with patch 1, patch 2, and patch 3 surfaces of GLD-2 (right, middle, and left panels, respectively). In the middle panel, note that the RNP-8GB helix is close to helix α4 of the GLD-2 central domain and that the GLD-2 loop containing Arg756 binds on top of the RNP-8GB helix. (C) Corresponding zoom-in of GLD-3NT interactions with the same surface patches of GLD-2 (PDB code 4ZRL, Nakel et al. 2015). In the left panel, two GLD-3 residues previously shown to contribute to RNA binding are indicated (Arg42, Lys43, Nakel et al. 2015). In the middle panel, note that the GLD-3NT C-terminal helix is further apart from helix α4 of the GLD-2 central domain and that the GLD-2 loop containing Arg756 binds in between. (D) Superposition of RNP-8 and GLD-3 from the corresponding GLD-2–bound structures. The residues in the DDYV and ENFY segments are indicated. (E) Coomassie-stained 17% SDS/PAGE of His-pull-down experiments of coexpressed wild-type (wt) and mutant His-GLD-2PAP with RNP-8GB. Total lysate control is shown on the left; pulled-down protein eluate is shown on the right. The experiment includes the disrupting GLD-2 R547E mutant (which targets a central residue at the interaction interface) and as a control the GLD-2 N629A mutant (that still supports RNP-8 binding). (*) GST-6xHis-Tag contamination.