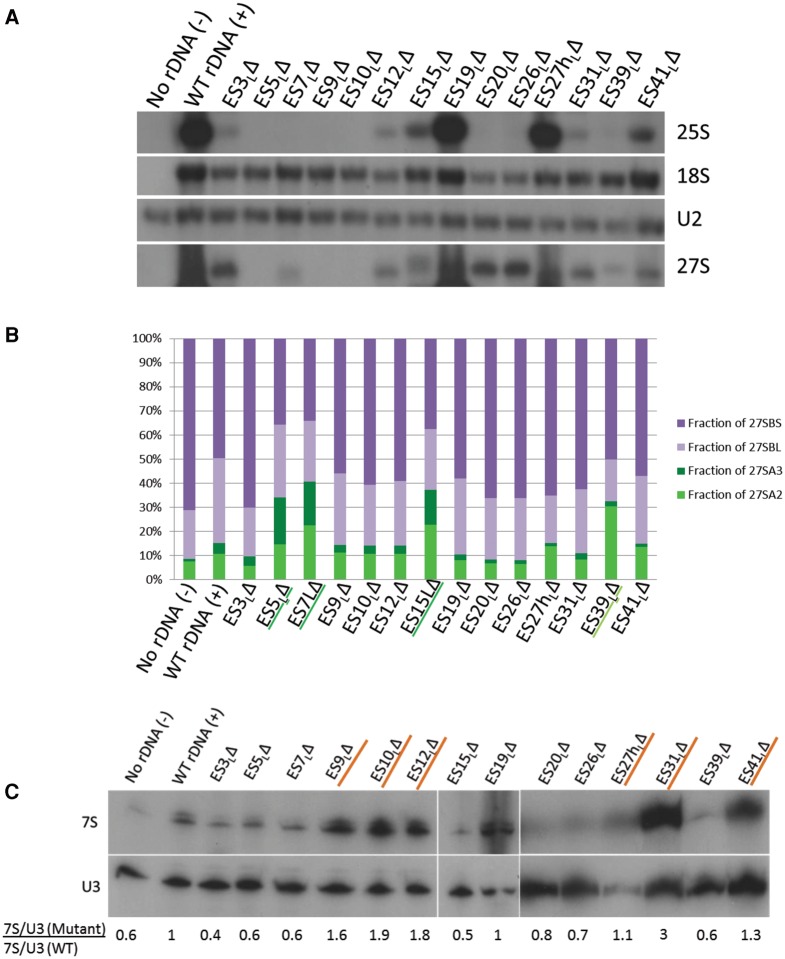
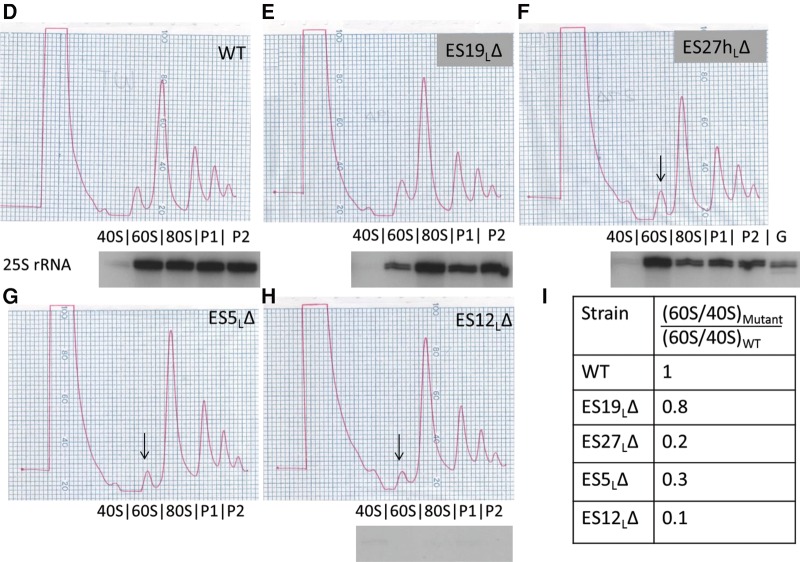
FIGURE 3.
Different ES participate in different steps of large ribosomal subunit assembly. (A) Representative Northern blot of total cellular RNA extracted from ES mutants, probed with various oligonucleotides to detect rRNAs and pre-rRNAs. Similar effects were observed in two different biological replicates. U2 snRNA is the loading control. Only the plasmid-derived 25S, 18S and 27S rRNAs are detected, since plasmid-specific oligonucleotide tags were used to probe for these rRNA species. (B) Relative steady-state levels of various 27S pre-rRNAs (27SA2, 27SA3, 27SBL, 27SBS) in ES mutants, assayed by primer extension. After normalizing each lane with respect to the U2 snRNA loading control, the fraction of each species of pre-rRNA (27SA2, 27SA3, 27SBL, or 27SBS) was calculated as a percentage of total 27S pre-rRNA in that lane (27SA2 + 27SA3 + 27SBL + 27SBS). 27SA processing intermediates are shown in shades of green (27SA2 is light green and 27SA3 is dark green). 27SB processing intermediates are shown in shades of purple (27SBL is light purple and 27SBS is dark purple). An increase in green 27SA pre-rRNA and a concomitant decrease in purple 27SB pre-rRNA are diagnostic of an “early” ES mutant. Note that the levels of pre-rRNA species observed here are a combination of both residual endogenous WT and plasmid-derived mutant pre-rRNA. Shown here are the mean values from three biological replicates. For more details and error bars, see Supplemental Figure 3. (C) Northern blot of total cellular RNA extracted from ES mutants, probed with oligonucleotides that hybridize to 7S pre-rRNA. U3 snoRNA is the loading control. Shown below the blot is the fold increase of 7S pre-rRNA in mutants compared to WT, normalized for loading. A mutant that has a ratio of >1 accumulates 7S pre-rRNA (characteristic of late-acting ES, highlighted in orange), and a mutant that has a ratio of <1 has decreased levels of 7S pre-rRNA (seen in early or middle phenotypes). (D–I) Sucrose gradient centrifugation to assay defects in 60S ribosomal subunit biogenesis in WT cells (D), viable ES mutants (E,F), and representative lethal ES mutants (G,H). Arrows shown in G, H, and F indicate the decreased 60S subunit peak in the lethal ES mutants and in the viable ES27hLΔ mutant, respectively, compared to WT (B). In this assay, defects in 60S subunit biogenesis are discerned exclusively by a decreased 60S peak and a decreased ratio of 60S to 40S subunits, compared to WT. Plasmid-derived 25S rRNA detected by reverse transcription using an oligonucleotide complementary to the plasmid-specific tag is shown underneath the polysome curves to indicate those fractions that contain plasmid-derived mature 25S rRNA. As shown in F, a sequencing lane with G was used to map the 5′ end of 25S rRNA. The primer extension assay could not be performed on the ES5LΔ mutant in G, since this deletion encompasses the plasmid-specific tag, thereby eliminating the binding site of the reverse transcription primer. (G) Ratios of area under the curve of 60S/40S subunits in mutant compared to WT, quantified from D–H. All the ES mutants shown, except ES19LΔ, have a multifold decrease in 60S/40S ratio compared to WT, indicative of defects in 60S subunit biogenesis.