Abstract
Trypanosoma brucei, the etiologic agent of sleeping sickness, encodes a single intron-containing tRNA, tRNATyr, and splicing is essential for its viability. In Archaea and Eukarya, tRNA splicing requires a series of enzymatic steps that begin with intron cleavage by a tRNA-splicing endonuclease and culminates with joining the resulting tRNA exons by a splicing tRNA ligase. Here we explored the function of TbTrl1, the T. brucei homolog of the yeast Trl1 tRNA ligase. We used a combination of RNA interference and molecular biology approaches to show that down-regulation of TbTrl1 expression leads to accumulation of intron-containing tRNATyr and a concomitant growth arrest at the G1 phase. These defects were efficiently rescued by expression of an “intronless” version of tRNATyr in the same RNAi cell line. Taken together, these experiments highlight the crucial importance of the TbTrl1 for tRNATyr maturation and viability, while revealing tRNA splicing as its only essential function.
Keywords: Trypanosoma, tRNA, tRNA editing, splicing, intron
INTRODUCTION
Transfer RNAs (tRNAs) are essential for maintenance of cellular homeostasis as central players for mRNA decoding during protein synthesis in the ribosome. The biosynthesis of mature, functional tRNAs requires several processing steps, including editing and post-transcriptional modification events, removal of 5′ leader and 3′ trailer sequences, addition of the 3′-terminus -CCA sequence; and when present, removal of introns by a specialized tRNA splicing machinery.
Introns have been described in tRNAs in the three kingdoms of life. In eukaryotes, the number of intron-containing tRNAs genes varies among different organisms, ranging from 17 tRNA isoacceptors in Danio rerio, three in Arabidopsis thaliana, 10 in Saccharomyces cerevisiae, and 10 in humans; each of these containing 813, 83, 61, and 34 intron-containing tRNA genes, respectively (Chan and Lowe 2009). In trypanosomatids, which include medically important pathogens of the genus Leishmania and Trypanosoma, tRNATyr is the only intron-containing tRNA isoacceptor; a single-copy gene in T. brucei but that may occur in multiple copies in other eukaryotes.
In addition, trypanosomatid tRNA introns are among the shortest known in biology, with sizes between 11–13 nucleotides (nt), which approaches the proposed theoretical minimum for splicing (Tocchini-Valentini et al. 1993). Despite these differences in tRNATyr copy number, intron length, and sequence content, the mature tRNATyr is still highly conserved and has identical sequences in the different trypanosomatids (Padilla-Mejía et al. 2009). However, because in all eukaryotes the trypanosomatid tRNA introns interrupt the anticodon loop, splicing is still an essential step in maturation and generation of a fully functional tRNATyr.
Intron removal in tRNA occurs by two different mechanisms: (i) In bacteria, tRNAs contain Group I introns (Reinhold-Hurek and Shub 1992), which are self-splicing and whereby intron removal occurs concomitantly with tRNA exon ligation; (ii) in Archaea and Eukarya, tRNA introns are removed by an all-protein tRNA endonuclease, which generates two tRNA halves with different termini; a 2′,3′-phosphate at the 5′-exon and a 5′-OH at the 3′-exon (Abelson et al. 1998). Ligation of the tRNA exons in Archaea and most Eukarya is catalyzed in the 3′–5′ direction (3′–5′ pathway) by the RtcB enzyme, using the 3′-phosphate left at the 5′-exon and the 5′-OH terminus of the 3′-exon (Filipowicz and Shatkin 1983; Popow et al. 2011). RtcB was first described in Escherichia coli as part of a two-gene operon with RNA cyclase (RtcA), an enzyme responsible for the conversion of RNA 3′-phosphate to 2′,3′-cyclic phosphates (Genschik et al. 1997, 1998). Although RtcB is not the enzyme responsible for tRNA splicing in bacteria, it was shown that the E. coli RtcB is able to repair a ribotoxin induced breakage in a tRNA-like stem–loop structure, as well as function in tRNA and HAC1 mRNA splicing in a Δtrl1 yeast strain lacking the endogenous tRNA ligase Trl1 (Tanaka and Shuman 2011; Tanaka et al. 2011a). Today we know that RtcB executes the end-joining step of the RNA repair/splicing systems, requiring GTP as an energy source and Mn2+ as a cofactor (Tanaka et al. 2011b; Chakravarty and Shuman 2012; Chakravarty et al. 2012). Homologs of RtcB are widely spread among Eukarya, Bacteria, and Archaea, including trypanosomatids (Genschik et al. 1998).
Although most Eukarya share a common tRNA ligation pathway, a different pathway has been described for yeast and plants (Greer et al. 1983; Phizicky et al. 1992; Englert and Beier 2005). In these cases, tRNA ligation is performed by the enzyme Trl1 in the 5′–3′ direction (5′–3′ pathway) using a 5′-phosphate from the 3′-exon and the 3′-OH at the 5′-exon. However, prior to ligation, the terminus of each exon generated by cleavage has to be processed by Trl1; a multifunctional enzyme containing a central kinase domain that phosphorylates the 5′-OH of the 3′-exon, a C-terminal domain that hydrolyzes the 2′,3′-cyclic phosphate generating a 2′-phosphate/3′-OH terminus, and finally, an N-terminal ligase domain that can join the two tRNA exons (Greer et al. 1983; Sawaya et al. 2003; Remus and Shuman 2013). As a last step in maturation, the dangling 2′-phosphate, derived from the 2′–3′ cyclic phosphate intermediate, is removed by a trans-acting phosphotransferase (Tpt1), generating a fully mature functional tRNA (Spinelli et al. 1997).
Although based on sequence alignments, trypanosomatid homologs of yeast Trl1 have been reported, and their role in tRNA splicing has not been formally established. In the present report, we show that down-regulation of Trypanosoma brucei Trl1 (TbTrl1) by RNAi caused cell-cycle arrest at G1 and eventual death, highlighting the essentiality of this protein in these organisms. Unexpectedly, the absence of TbTrl1 leads to accumulation of intron-containing tRNAs, suggesting a level of coordination between cleavage and ligation during splicing. Furthermore, the expression of a mature version of tRNATyr, lacking the intron, rescues the growth defect caused by TbTrl1 down-regulation. Taken together, these data support the idea that TbTrl1 is an essential enzyme in T. brucei with an indispensable role in tRNATyr maturation.
RESULTS
T. brucei encodes a homolog of the yeast splicing ligase Trl1
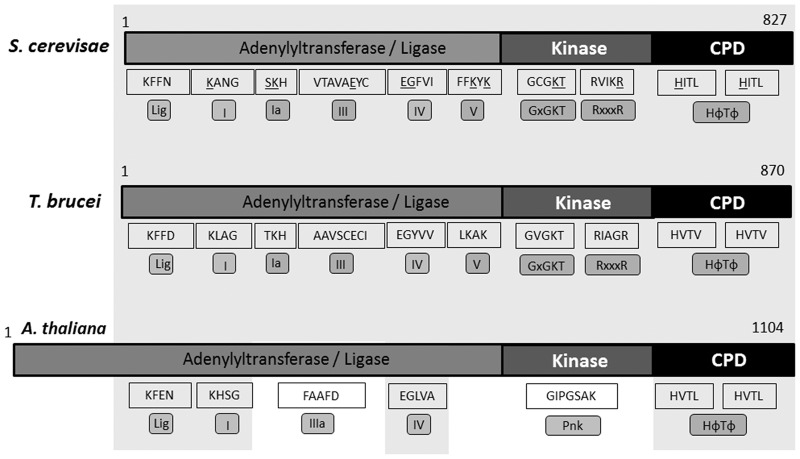
Potential homologs of yeast Trl1 from T. brucei, T. cruzi, and L. major were previously reported (Wang and Shuman 2005). Although overall these show sequence divergence when compared to the yeast and Arabidopsis Trl1, closer inspection revealed the presence of key essential residues that constitute the signature motif for 5′–3′-tRNA ligases (Fig. 1; Wang et al. 2002; Sawaya et al. 2003). A search for sequence motifs in the putative TbTrl1 enzyme using the NCBI conserved domain database shows the presence of a conserved tRNA splicing ligase domain. It also revealed a predicted P-loop sequence harboring a conserved nucleotide phosphate-binding motif found in many kinases (Marchler-Bauer et al. 2015). The presence of the Trl1 N-terminal adenylyltransferase/ligase domain is also evident, including five different conserved domains described as I, Ia, III, IV, and V, showing the signature sequences KLAG, TKH, AAVSCECI, EGYVV, and LKAK, respectively. These motifs include essential residues for activity such as the conserved lysines (K) in motifs I, Ia, and V; glutamic acids (E) in III and IV; and glycine (G) in motif IV (Sawaya et al. 2003). The central kinase domain harbors the key motifs GxGKT and RxxxR at positions 400–404 and 524–528 of the predicted T. brucei protein sequence, respectively. Finally, the C′-terminal cyclic phosphodiesterase (CPD) domain contains the two conserved HVTV repeats (HФTФ motifs) at positions 739–742 and 802–805 (Fig. 1).
FIGURE 1.
Schematic representation of Trl1 sequences. Trl1 sequences from Saccharomyces cerevisiae (827 aminoacids), Trypanosoma brucei (870 amino acids), and Arabidopsis thaliana (1104 amino acids) were analyzed. The gray area shows conserved regions and motifs between the sequences. Motifs Lig, I, Ia, III, IIIa, IV, and V are present in the ligase domain. Motifs GxGKT, RxxxR, and Pnk are conserved in the kinase domain. Motif IIIa and Pnk are observed only in plants. The CPD domain is marked by the presence of two HФTФ motifs. All the essential amino acid residues in yeast Trl1 ligase are underlined.
TbTrl1 is essential in T. brucei
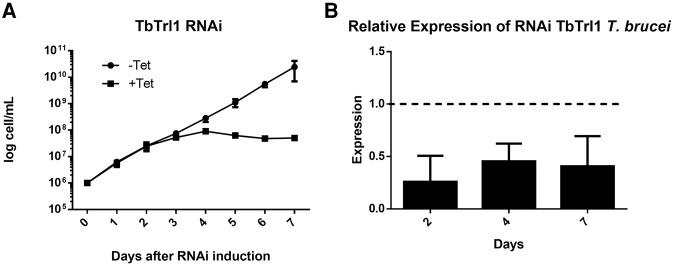
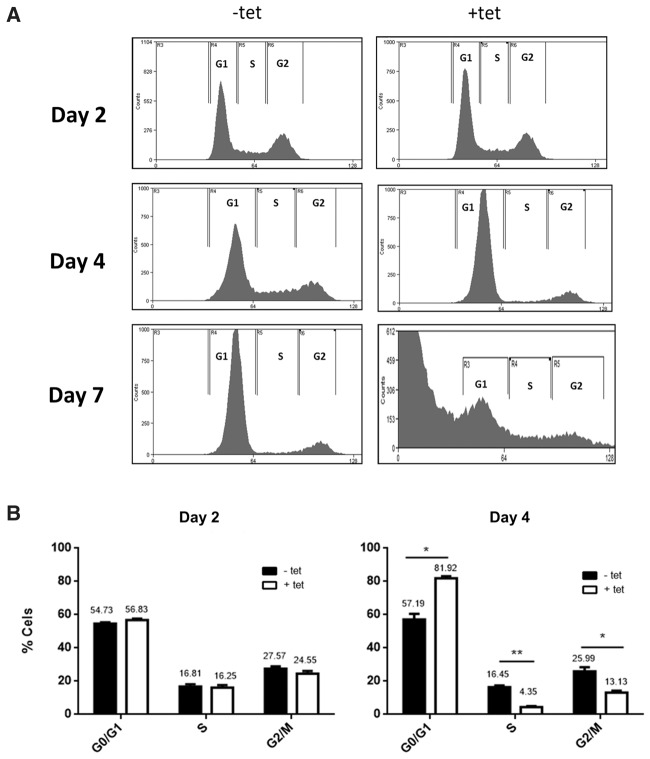
To explore the potential function of TbTrl1, a tetracycline inducible T. brucei RNAi strain was generated for TbTrl1 gene silencing. Growth curves of RNAi-induced and uninduced cells were performed, revealing that down-regulation of TbTrl1 leads to slow growth after 3 d of induction (Fig. 2A). Down-regulation of TbTrl1 was confirmed by qPCR showing a 75% reduction in TbTrl1 mRNA levels (Fig. 2B). To determine whether the growth inhibition was attributable to cell-cycle arrest and/or cell death, flow cytometric analysis was performed. After 2 d of RNAi induction, there were no differences in the cell-cycle profile of induced and uninduced cells. However, upon 4 d of TbTrl1 down-regulation, 25% of the cells get arrested at the G1 phase of the cell cycle (Fig. 3A). G1 arrest was accompanied by a decrease in the number of cells in S (compare 4% for silenced cells to 16% for nonsilenced) and G2 phases (13% for silenced and 26% for nonsilenced) (Fig. 3B). After 7 d of RNAi induction, the majority of cells were in the sub G0 phase, which precedes the G1 peak and is suggestive of DNA degradation and thus, cell death. These results show that TbTrl1 is essential for the T. brucei viability and its reduced expression compromises the cell-cycle progression of the parasites, leading to cell death.
FIGURE 2.
Effects of TbTrl1 RNAi on the growth rate of procyclic form T. brucei cells. (A) Cloned procyclic trypanosome cells harboring the TbTrl1 RNAi plasmid construct were incubated in culture medium without (−tet, black circles) or with (+tet, black squares) tetracycline. The rate of cell growth was monitored daily, and the cell numbers were plotted in a logarithmic scale as cumulative cell numbers. (B) tbtrl1 intracellular mRNA levels monitored by qPCR from the corresponding RNAi experiments after 2, 4, and 7 d of induction. The expression levels were normalized using α-tubulin as an endogenous control, P < 0.05 (n = 3).
FIGURE 3.
FACS analysis of the TbTrl1-deficient procyclic form T. brucei cells. Samples of procyclic form T. brucei cells harboring the TbTrl1 RNAi plasmid construct were taken at various times for FACS analysis following tetracycline induction. (A) Histograms from FACScan of induced (+tet) and uninduced cells (−tet). (B) Percentages of induced (+tet) and uninduced cells (−tet) in G1, S, and G2/M phases as determined by the WinMDI software.
TbTrl1 is essential for tRNATyr maturation in T. brucei
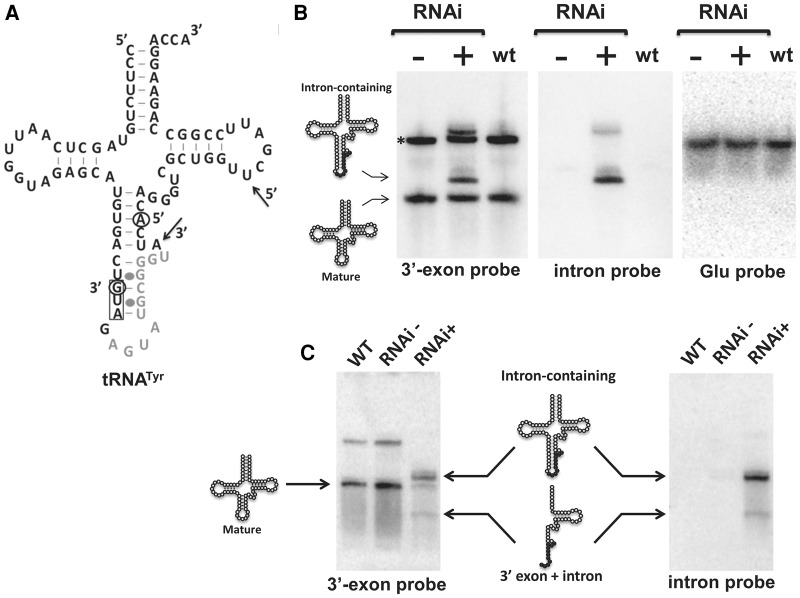
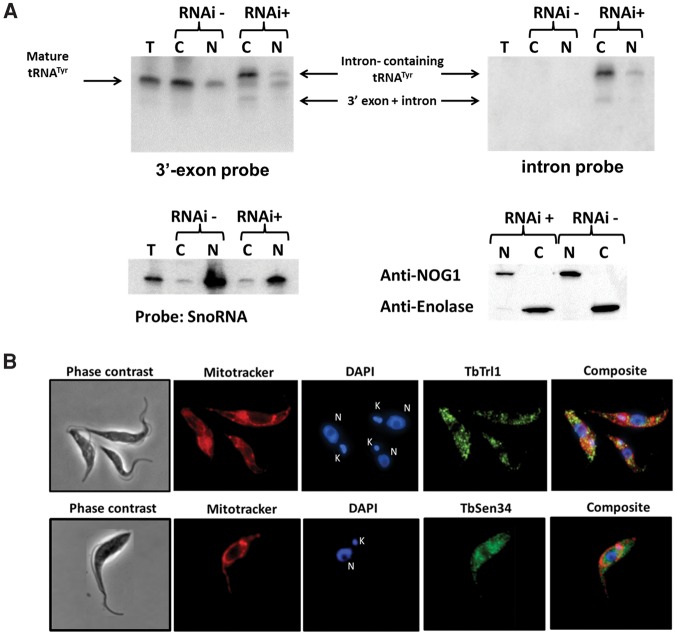
To evaluate TbTrl1 function in tRNATyr maturation, total RNA was collected from 2- and 4-d-induced and uninduced cells and analyzed by Northern blots with oligonucleotide probes specific for tRNATyr (Fig. 4A). Two days after RNAi induction, we observed accumulation of intron-containing tRNATyr (Fig. 4B), an unexpected result, given that down-regulation of ligase function should lead to accumulation of unligated exon halves. However, we note that 2 d represent a time point early in the RNAi induction, even before the appearance of the growth defect. After 4 d of down-regulation of TbTrl1 expression, a tRNATyr fragment was detected with the 3′exon probe. An identical migrating band was also observed with the intron-specific probe, indicating the presence of a tRNATyr 3′exon–intron intermediate (Figs. 4C, 6A). We performed similar experiments with several probes specific for the 5′exon but unfortunately were unable to detect 5-exon accumulation, or for that matter, mature tRNA. We suspect there may be post-transcriptional modifications on the 5′half of the tRNA that hinder probe hybridization. Regardless, after 4 d of RNAi induction, there is also a commensurate reduction in the levels of mature tRNA when compared to the nonsilenced control and also with wild-type T. brucei (Fig. 4C), consistent with a defect in tRNA splicing caused by down-regulation of TbTrl1. To verify the possibility that the observed defects were splicing specific, we also probed for the non-intron containing tRNAGlu, which showed no difference in levels regardless of the cell line (Fig. 4C). In all conditions tested we also observed the presence of a larger product (Fig. 4B, asterisk). Currently, we do not know the significance of this larger product but assume that this is the 3′-extended pre-tRNATyr previously described (Schneider et al. 1993). Regardless, taken together these data strongly support that the lack of TbTrl1 specifically affects tRNATyr maturation. Importantly, the accumulation of intron-containing tRNATyr suggests connectivity between exon ligation and intron cleavage.
FIGURE 4.
Effects of TbTrl1 knockdown on tRNATyr maturation. (A) The cloverleaf structure of the tRNATyr of T. brucei. The 11-nt intron is shown in gray letters. The anticodon sequence is boxed. Black arrows and open circles denote the positions where the 3′-exon Tyr probe and Intron probe anneal, respectively (5′ and 3′ nucleotides are marked). (B, C) Northern blot analysis of tRNATyr processing steps in wild-type (WT) and TbTrl1 RNAi strain induced (+tet) and uninduced (−tet) with tetracycline after 2 d (B) or 4 d (C) of RNAi. The positions of the intron-containing, mature tRNATyr and 3′-exon–intron species signals are indicated. The asterisk (*) in panel B indicates a probable 3′-extended pre-tRNATyr previously described (Schneider et al. 1993). A tRNAGlu probe was used as control (B).
FIGURE 6.
Subcellular localization of tRNATyr, TbTrl1, and TbSen34. (A) RNA from total (T), nuclear (N), and cytoplasmic (C) fraction of 4 d induced (+) and uninduced (−) TbTrl1 RNAi cells was extracted and analyzed by Northern blot using a 3′ exon or intron probe. The intron-containing, mature tRNATyr and the 3′-exon–intron species are indicated by arrows. SnoRNA probe was used as control for nuclear fraction purity. The purity of the fractions was confirmed by Western blotting using anti-NOG1 (specific for nucleus) and anti-enolase antibodies (specific for cytoplasm). (B) Immunolocalization of His-tag TbTrl1 and Protein C-TbSen34. Anti-His-Tag and Anti-Protein C were used as primary antibodies for His-Tag TbTrl1 and detection Protein C-TbSen34, respectively. MitoTracker red was used as a mitochondrial marker, while DAPI was used to mark the position of nuclear (N) and mitochondrial DNA (kinetoplast) (K). TbTrl1 and TbSen34 are shown in green. A background was added to the TbSen34 images in order to get a more harmonious figure.
tRNATyr maturation is the only essential function of TbTrl1
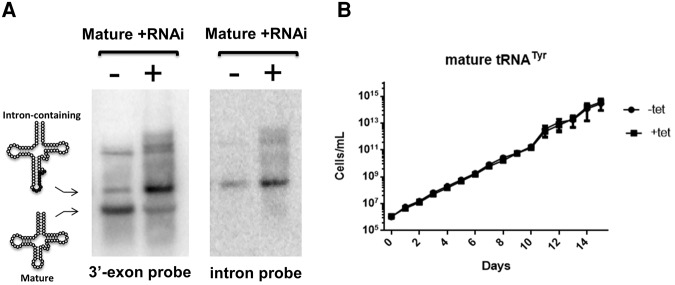
Since tRNATyr is the product of a single-copy gene, its proper maturation is essential for protein synthesis. The fact that intron-containing tRNA accumulates upon TbTrl1 RNAi induction suggests that either the growth defect, and implicitly the G1 arrest, is the result of reduced levels of mature tRNATyr (Fig. 4B) or alternatively it is the result of accumulation of intron-containing tRNA somehow serving as a signal for arrest. Unspliced tRNA rapidly accumulates in the nuclei of yeast S. cerevisiae after DNA damage. This provided evidence for signal-mediated crosstalk among tRNA nucleocytoplasmic trafficking, protein synthesis, and checkpoint execution, enabling functional coupling of tRNA biogenesis and cell-cycle progression (Ghavidel et al. 2007). To differentiate between these possibilities, we coexpressed a mature copy of tRNATyr (lacking the intron) in the TbTrl1 RNAi cell line and performed similar growth curves while comparing RNAi-induced and uninduced cells. We found that mature tRNATyr expression rescued the growth defect of the TbTrl1 RNAi strain (Fig. 5B). Northern blots of total RNA isolated from the “rescued” strain after 4 d of tetracycline induction showed that intron-containing tRNA still accumulated, but in addition, the complementing mature tRNATyr was successfully expressed (Fig. 5A). Despite the fact that the relatively mature tRNA expression was lower than the endogenous intron-containing tRNA resulting from RNAi, it was still sufficient to rescue the growth defect (Fig. 5). Similar rescue was not observed when a nonrelated tRNA was expressed as a control under similar RNAi conditions (data not shown). These data strongly support the idea that the only essential function of TbTrl1 is to ensure proper tRNATyr maturation, which in turn is essential for cell growth.
FIGURE 5.
Effects of mature tRNATyr expression on the TbTrl1-deficient procyclic form T. brucei cells. (A) Northern blot analysis of 4 d RNAi induced (+tet) and uninduced TbTrl1 RNAi cells expressing recombinant mature tRNATyr. Specific probes were used for Intron-containing (Intron probe) and mature (3′ exon Tyr probe) tRNATyr. The position of the Intron-containing and mature tRNATyr signals are indicated on the left. (B) Growth curve of uninduced (−tet, black circles) and induced (+tet, black squares) TbTrl1 RNAi expressing mature tRNATyr. The y-axis shows a log-scale of cumulative cell densities accounting for total dilutions and the x-axis shows the progression of the growth curve in days.
Cytoplasmic localization of tRNA splicing components in T. brucei
In yeast, the tRNA splicing endonuclease is found on the surface of the outer mitochondrial membrane facing the cytoplasm; the tRNA ligase is also cytoplasmic (Yoshihisa et al. 2003). However, the subcellular localization of tRNA splicing components varies between different organisms. In Xenopus oocytes, all the components of the tRNA endonuclease complex, as well as the tRNA ligase, are localized to the nucleus (De Robertis et al. 1981). In plants, the tRNA endonuclease localizes both to the nucleus and the mitochondria and the tRNA ligase was predominantly targeted to chloroplasts (Englert et al. 2007). To determine where tRNA splicing localizes in T. brucei, we performed Northern blots with a radioactively labeled oligonucleotide probe specific for the 3′ exon portion of the tRNA. Notably, this probe does not discriminate between mature and unspliced tRNA; however, these two species can be easily separated by electrophoresis. We used nuclear and cytosolic fractions of the TbTrl1 RNAi silenced parasites and compared these to similar fractions from uninduced cells. We observed accumulation of intron-containing tRNATyr in the cytoplasm, while equivalent amounts of mature and unspliced tRNA were seen in the nuclear fraction (Fig. 6A). In addition, smaller fragments corresponding to tRNATyr 3′exon–intron intermediates, predictably generated by splicing endonuclease cleavage prior to ligation, are present only in the cytoplasmic fraction (Fig. 6A). These results suggest that tRNA splicing occurs in the cytoplasm of T. brucei. To rule out the possibility of cross-compartment contamination, the same RNA fractions were probed for a nucleolar marker (snoRNA 82). In this control experiment, clear and substantial enrichment of the snoRNA marker was observed in the nuclear fractions. Furthermore, we also used protein samples derived from the same cell fractionation experiment for Western analysis, to further test for cross-contamination. We used antibodies to compartment specific markers NOG1 (nucleolar) and enolase (cytoplasmic). This experiment again revealed negligible cross-contamination of our intracellular fractions. We also performed localization experiments with cells expressing an epitope-tagged version of the tRNA ligase and TbSen34, a catalytic subunit of T. brucei tRNA endonuclease. For this purpose, two different T. brucei strains harboring a plasmid for tetracycline-inducible expression of the TbTrl1 protein with a C-terminus His-tag or TbSen34 with an N-terminus protein C were generated. After induction of each TbTrl1 and TbSen34 expression with tetracycline, the localization of each protein was individually determined in each strain by immunofluorescence using anti-His tag or anti-protein C antibody, respectively. In both strains we observed fluorescent punctate signals scattered in the cytoplasm (Fig. 6B), suggesting again that pre-tRNATyr splicing takes place in the cytoplasm of T. brucei procyclic cells. Taken together, these experiments strongly support the idea of cytoplasmic localization of tRNA splicing in T. brucei.
DISCUSSION
Splicing is an essential step for tRNA maturation in all eukaryotes. Although the intron removal step is catalyzed by a highly conserved endonuclease in Archaea and Eukarya, the final ligation step occurs by two different mechanisms. The 3′–5′ ligation pathway involves the RtcB protein, which is found in Bacteria, Archaea, and Eukarya (Genschik et al. 1998). The 5′–3′ ligation pathway is catalyzed by a tRNA ligase with a very restricted phylogenetic distribution, which is described in plants, yeast, and trypanosomatids. Interestingly, trypanosomatids, together with Branschiostoma floridae, are the only organisms known to encode both RtcB and Trl1 homologs (Englert et al. 2010). Notwithstanding the existence of RtcB in trypanosomes, our data strongly point to TbTrl1 as the tRNA splicing ligase in T. brucei. This is supported by the observed accumulation of tRNATyr processing intermediates when expression of TbTrl1 is down-regulated. As for TbRtcB, its function is not currently clear in T. brucei; however, eubacterial genomes harbor RtcB genes despite encoding self-splicing intron-containing tRNAs (Kuhsel et al. 1990; Reinhold and Shub 1992; Biniszkiewicz et al. 1994). In these cases, RtcB is involved in RNA repair as previously proposed for E. coli (Tanaka and Schuman 2011) and the same may be true of the T. brucei homolog.
The reduced levels of mature tRNATyr and the accumulation of the pre-tRNA species are in line with the observed growth defects. In E. coli, reduced levels of functional tRNAs interfere with the rate of protein synthesis with concomitant growth retardation and cell death (Mohanty and Kushner 2013). In yeast, the accumulation of intron-containing tRNA results in G1 cell-cycle arrest by Gcn4 activation and Cln2 accumulation (Ghavidel et al. 2007), which is tantamount to the effect we observed in the parasite after Trl1 silencing. However, the signal that leads to cell-cycle arrest in T. brucei is not due to accumulation of unspliced tRNATyr, as expression of mature tRNATyr was able to rescue the growth defects, despite the presence of significant levels of unspliced tRNA in the RNAi-induced TbTrl1 strain (Fig. 5). In addition, trypanosomatids lack obvious homologs of yeast Gcn4, suggesting a different signaling pathway for cell-cycle arrest in response to tRNA impairment. Nonetheless, we cannot formally discard the possibility that effects on mature tRNA levels will reflect onto changes in protein synthesis rates, potentially driving the signaling for cell-cycle arrest. In fact, the Kluyveromyces lactis zymocin, which cleaves the anticodon loop of modified tRNA, leads to cell-cycle arrest in G1 (Butler et al. 1991; Lu et al. 2005). Killer toxins from Pichia acaciae (PaT) and Wingea robertsiae, responsible for cleavage, provoke S-phase arrest and concomitant DNA damage checkpoint activation (Klassen and Meinhardt 2005; Klassen et al. 2004, 2008).
In a similar manner, the E. coli anticodon nuclease PrrC (EcoPrrC) leads to S. cerevisiae growth arrest; in this latter case, growth can be resumed by expression of the toxin target, tRNALys (Meineke et al. 2011). As discussed above, PaT targets a specific tRNA in a mode of action that resembles that of zymocin, but it arrests the cell cycle in the S phase, not G1. This cell-cycle arrest is presumably mediated by Rad53p checkpoint phosphorylation. The mechanisms that lead to cell death or growth arrest after tRNA cleavage are not well understood and seem to vary for the different ribotoxins. A recent study showed that PaT Orf2p (the toxic subunit) cleaves DNA in addition to its specific tRNA cleavage activity, and translation impairment caused by tRNA cleavage was responsible for the Orf2p relocation to the nucleus, leading to histone phosphorylation and cell-cycle arrest (Shigematsu et al. 2013). In S. cerevisiae, diverse cell-cycle phenotypes have been described after mutations leading to loss of function in genes associated with the protein synthesis machinery (Polymenis and Aramayo 2015). It is then possible that damage in protein synthesis leads to nucleus relocalization of an unknown factor that is responsible for arrest of cell-cycle progression in T. brucei. In vitro competition assays using yeast tRNA endonuclease, Trl1, T4 RNA ligase and kinase, show that excision and joining of tRNA halves are concerted events and provide evidence for the assembly of a functional tRNA splicing complex in yeast (Greer 1986). In humans, the association of tRNA splicing enzymes has already been demonstrated and defects in Sen2, one of the endonuclease subunits, impairs pre-tRNA maturation (Paushkin et al. 2004; Popow et al. 2011). Our data are in agreement with the observations above, indicating the tRNA endonuclease reaction cannot proceed efficiently given the accumulation of pre-tRNATyr and the 3′-exon–intron species in the absence of the tRNA ligase. More evidence supporting the existence of a tRNA splicing complex in T. brucei is the presence of both TbTrl1 and tRNA endonuclease Sen34 subunit (this work) in cytoplasmic granules, although colocalization of the two enzymes is still to be confirmed. The subcellular distribution of the tRNA splicing machinery components is extremely variable in different organisms. The yeast endonuclease is attached to the outer membrane of the mitochondria; ScTrl1 is distributed in the cytoplasm, where the splicing event occurs (Yoshihisa et al. 2003). Plant tRNA ligases show a different subcellular localization, partitioning to chloroplasts (Englert et al. 2007).
The retention of a single intron-containing tRNA in T. brucei is intriguing. In other organisms, the isomerization of the U35 to pseudouridine in tRNATyr has been shown to depend on the intron presence (Choffat et al. 1988; van Tol and Beier 1988; Pieńkowska et al. 2002). A yeast knockout strain for the enzyme responsible for this modification, Pseudouridine synthetase 7, presents higher sensitivity to some drugs and heat shock, but the cells are still viable under the growth conditions used (Sinha et al. 2008; Westmoreland et al. 2009). In T. brucei, while we have not shown the presence or absence of the pseudouridine at position 35 of tRNATyr, clearly the mature tRNA expressed without the intron is able to rescue the TbTrl1 RNAi strain; therefore, while intron-dependent pseudouridine may still be important, it is not at the root of the growth defect when TbTrl1 is down-regulated and likely not the reason for intron retention. Previous work showed the presence of several noncanonical editing events in the tRNATyr intron and absence of anticodon modifications in the pre-tRNA. Editing is required for proper intron processing by the tRNA endonuclease (Rubio et al. 2013), adding another level of complexity to the maintenance of a single intron. As suggested by Rubio and coworkers, the intron may ensure proper L-shape folding of the tRNA, with the editing events providing order to the folding process and avoiding the establishment of undesirable mature tRNA conformations. Another possibility is that certain life threatening conditions, such as amino acid starvation and DNA damage, might dictate the existence of a salvage pathway. The balance of nuclear and cytoplasmic mature tRNA pools, resulting from proper/nonproper function of the tRNA splicing pathway, can be responsible for triggering a transitory signal for cell-cycle arrest. In agreement with this hypothesis, our results show that while mature tRNA levels are decreased, we see accumulation of intron-containing tRNA in the cytoplasm and both species are present in the nucleus.
MATERIALS AND METHODS
Cell cultivation and cell quantitation
Procyclic forms of T. brucei strain 29-13 were grown at 28°C in SDM-79 media containing hygromycin (50 µg/mL) and G418 (15 µg/mL) and supplemented with 10% heat-inactivated bovine fetal serum. TbTrl1 RNAi strain and TbTrl1 RNAi expressing mature tRNATyr were grown in the same media containing phleomycin (2,5 µg/mL) and phleomycin (2.5 µg/mL) plus puromycin (1 µg/mL), respectively. The number of cells was determined by counting using a Neubauer's chamber.
Plasmids construction, transfection, and RNAi induction
To create the TbTrl1 RNAi strain, a dual hairpin RNAi plasmid with a tetracycline-inducible promoter containing a 600-bp fragment of the TbTrl1 gene (from position 720 to position 1320) (Tb10.6k15.2400) was constructed using the vectors pLEW100 and pJM325 as previously described (Shi et al. 2000; Wang et al. 2000). The TbTrl1 gene fragment was amplified from genomic DNA by PCR using the oligonucleotides trl1F: 5′-AAGCTTACGCGTGAGAGGCAGCATTGGGAAGGGAAAGATCC-3′ e trl1R: 5′-TCTAGAATGGTGCTCCGCCACGCACTTCGCA-3 and the High-Fidelity mix from Thermo Scientific. The final plasmid was linearized with EcoRV before transfection in T. brucei for genomic integration. Procyclic forms were transfected using AMAXA Nucleofector II (Lonza Group AG) with Program X-001 and Human T-cell Nucleofector solution. The T. brucei clones were obtained by limiting dilution in phleomycin-contained media and the TbTrl1 RNAi was induced with 1 μg/mL of tetracycline.
The mature tRNATyr expression plasmid was created from pEA-5 (gift from Dr. Andre Schneider, University of Bern, Switzerland). A 592-nt fragment, including the intronless-tRNATyr and its promoter, was synthesized by Genescript and it was cloned into pEA-5 using KpnI and XhoI restrictions sites as previously described in Aeby et al. (2010).
To express TbTrl1, the C′-terminal tagged TbTrl1 protein was produced by PCR amplification of the full-size TbTrl1 gene and cloned into the pLEW79-MHTAP plasmid containing His and Myc epitope tags (Jensen et al. 2007).
Cell-cycle analysis
Time samples with 1 million cells of the RNAi plasmid transfected T. brucei 29-13 strain were collected before and during tetracycline induction of gene knockdown. Cells were initially washed with PBS and then incubated in PBS containing 0.1% Triton X-100, 10 µg/mL RNAse, and 10 µg/mL propidium iodide. Subsequently, they were incubated at 25°C for 10 min and analyzed by flow cytometry. One hundred thousand cells were examined using the FACScan system flow cytometer (Becton, Dickinson and Company). Percentage of cells in each phase of the cell cycle, G1, S, and G2/M, was determined using the WinMDI software (Multiple Document Interface Flow Cytometry Application software, V2.8 (http://facs.scripps.edu/software.html).
Northern blot analysis
Total RNA or nuclear RNA from induced and noninduced cells was isolated using the guanidinium thiocyanate/phenol/chloroform extraction method (Chomczynski and Sacchi 1987). Briefly, 15 µg of total RNA was separated on denaturating 8% polyacrylamide gel with 8 M urea and electroblotted to Zetaprobe (Bio-Rad) membranes. The membrane was incubated in a prehybridization solution (5× SSC; 20 mM sodium phosphate pH 7.2; 7% SDS; 1× Denhardt's solution) containing 10 µg/mL of ssDNA for 30 min. The membranes were subsequently probed with [32P]-5′-end labeled oligonucleotides specific for the 3′-tRNATyr exon (5′-AACCAGCGACCCTGTGAT-3′), tRNATyr intron (5′-TGATACCTGCAAACTCTAC-3′), (5′-TTCCGGTACCGGGAATCGAAC-3′), or snoRNA82 (5′-CAACGTCCATCTGCGACGGCTTTA-3′) for 18 h at 42°C and washed. Images were recorded with a Storm Phosphor imager (Bio-Rad).
Nuclear preparation
T. brucei cells in exponential growth (5 × 106 cells/mL) were harvested by centrifugation (2000g for 20 min) and washed with phosphate-buffered saline. About 8 × 108 cells/mL were suspended in lysis buffer (1 mM PIPES pH 7.4, 0.5 M hexylene glycol, and 1 mM CaCl2) and broken using a Stanstedt power fluid apparatus at 20 ψ (Shapiro and Doxsey 1982). The cytosolic fraction was obtained by centrifugation at 2500g for 20 min. The pellet was resuspended in lysis buffer and loaded in a 35% Percoll gradient for nuclei isolation (Amersham Pharmacia Biotech). The gradient was created by centrifugation at 60,000g for 35 min and the nuclei were collected by a side puncture. After collection, the nuclei were washed in 1 mM PIPES pH 7.4, 0.5 M hexylene glycol, 1 mM CaCl2, and 0.75 M sucrose and isolated by centrifugation at 2000g for 20 min. Following gradient purification, RNAs were extracted by the guanidinium method (Chomczynski and Sacchi 1987).
Immunolocalization and Western analysis
The protein-coding region of TbTrl1 was cloned into the T. brucei protein expression vector pLEW79-MHTAP, which places a His-tag at the C-terminus of the protein. For TbSen34, the protein-coding region of the gene was fused to a protein C tagged at its N-terminus and cloned into the pLEW83 T. brucei expression plasmid. The constructs were transfected into procyclic T. brucei 29-13 cells and clonal cell lines were obtained by limiting dilution. TbTrl1 and TbSen34 expression was induced in each strain by the addition of tetracycline for 24 h and intact cells were fixed and then stained with 200 nM MitoTracker Red dye (Life Technologies) for 20 min at 27°C, followed by washing and fixing to slides with 4% paraformaldehyde. Mitochondrial and genomic DNA were stained with DAPI shortly before immunofluorescence microscopy. The TbTrl1 localization was detected using an anti-His Tag antibody. The TbSen34 localization was detected using an anti-protein C antibody. Alexa Fluor 488-conjugated goat anti-mouse antibodies (Life Technologies) were used as the secondary antibodies. MitoTracker Red was added to the culture at 500 nM and shaken at 27°C for 30 min to allow uptake. Cells were harvested for microscopy at 2–5 × 106 cells/mL and washed twice in PBS before suspending in 3.7% formaldehyde/PBS solution, and 100 µL of suspension was fixed to glass slides for 10 min. Cells were washed three times in PBS, permeabilized with 0.1% Triton X-100 and washed an additional three times in PBS. Blocking was performed for 60 min using 5.5% FBS supplemented with 0.05% Tween 20 in PBS. All incubation steps were performed in a humid chamber and all antibodies were used at concentrations indicated in text. Cells were air-dried and mounted using Vectashield before imaging was performed using a Nikon Ti microscope. Image analysis was performed using ImageJ (NIH) and Nikon Elements AR.
Western blots were also used to determine the purity of sub-cellular fractions. For this, cell lysates from 5 × 106 cell equivalents of each fraction (nuclear or cytoplasmic) were loaded per well of a 15% SDS–polyacrylamide gel, blotted, and probed. The polyclonal rabbit antibodies against NOG1 and enolase (kindly provided by M. Parsons and P.A.M. Michels, respectively) were used at 1:100, 1:1.000, 1:1000, and 1:1.000 dilutions, respectively. Secondary anti-rabbit IgG antibodies (1:6000) coupled to horseradish peroxidase (GE Healthcare) were visualized according to the manufacturer's protocol using the ECL kit (Pierce).
ACKNOWLEDGMENTS
We thank Dr. Otávio Thiemann and Dr. Fernanda Costa for technical assistance with the RNAi experiments, Dr. E. Ullu and Dr. C. Tschudi for the RNAi plasmids pLEW100 and pJM325, Dr. A. Schneider for the tRNA expression plasmid, Dr. M. Parsons for the anti-NOG1 antibodies, Dr. P.A.M. Michels for the anti-enolase antibodies, and Dr. Vivian Rumjanek for assistance with the Flow Cytometry analysis. We are particularly grateful to Dr. Markus Englert for enlightened discussions. We thank Dr. Stewart Shuman and Dr. Najib El-Sayed for support and Dr. Dieter Söll and Dr. Claudio Masuda for helpful suggestions. The work was supported by the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação Carlos Chagas Filho de Amparo a Pesquisa de Estado do Rio de Janeiro (FAPERJ) with the aid of a National Institutes of Health GM084065-07 grant to J.D.A. and A.K. and a Czech Science Foundation (15-21450Y) grant to Z.P.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.056242.116.
REFERENCES
- Abelson J, Trotta CR, Li H. 1998. tRNA splicing. J Biol Chem 273: 12685–12688. [DOI] [PubMed] [Google Scholar]
- Aeby E, Ullu E, Yepiskoposyan H, Schimanski B, Roditi I, Mühlemann O, Schneider A. 2010. tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans. Nucleic Acids Res 38: 5833–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biniszkiewicz D, Cesnaviciene E, Shub DA. 1994. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J 13: 4629–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AR, White JH, Stark MJ. 1991. Analysis of the response of Saccharomyces cerevisiae cells to Kluyveromyces lactis toxin. J Gen Microbiol 137: 1749–1757. [DOI] [PubMed] [Google Scholar]
- Chakravarty AK, Shuman S. 2012. The sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res 40: 8558–8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Subbotin R, Chait BT, Shuman S. 2012. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci 109: 6072–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choffat Y, Suter B, Behra R, Kubli E. 1988. Pseudouridine modification in the tRNA (Tyr) anticodon is dependent on the presence, but independent of the size and sequence, of the intron in eucaryotic tRNA (Tyr) genes. Mol Cell Biol 8: 3332–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Black P, Nishikura K. 1981. Intranuclear location of the tRNA splicing enzymes. Cell 23: 89–93. [DOI] [PubMed] [Google Scholar]
- Englert M, Beier H. 2005. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res 33: 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Latz A, Becker D, Gimple O, Beier H, Akama K. 2007. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie 89: 1351–1365. [DOI] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Gundllapalli S, Beier H, Söll D. 2010. Branchiostoma floridae has separate healing and sealing enzymes for 5′-phosphate RNA ligation. Proc Natl Acad Sci 107: 16834–16839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Shatkin AJ. 1983. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell 32: 547–557. [DOI] [PubMed] [Google Scholar]
- Genschik P, Billy E, Swianiewicz M, Filipowicz W. 1997. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria and Archaea. EMBO J 16: 2955–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Drabikowski K, Filipowicz W. 1998. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its σ54-regulated operon. J Biol Chem 273: 25516–25526. [DOI] [PubMed] [Google Scholar]
- Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A. 2007. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell 131: 915–926. [DOI] [PubMed] [Google Scholar]
- Greer CL. 1986. Assembly of a tRNA splicing complex: evidence for concerted excision and joining steps in splicing in vitro. Mol Cell Biol 6: 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer CL, Peebles CL, Gegenheimer P, Abelson J. 1983. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell 32: 537–546. [DOI] [PubMed] [Google Scholar]
- Jensen BC, Kifer CT, Brekken DL, Randall AC, Wang Q, Drees BL, Parsons M. 2007. Characterization of protein kinase CK2 from Trypanosoma brucei. Mol Biochem Parasitol 151: 28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Meinhardt F. 2005. Induction of DNA damage and apoptosis in Saccharomyces cerevisiae by a yeast killer toxin. Cell Microbiol 7: 393–401. [DOI] [PubMed] [Google Scholar]
- Klassen R, Teichert S, Meinhardt F. 2004. Novel yeast killer toxins provoke S-phase arrest and DNA damage checkpoint activation. Mol Microbiol 53: 263–273. [DOI] [PubMed] [Google Scholar]
- Klassen R, Paluszynski JP, Wemhoff S, Pfeiffer A, Fricke J, Meinhardt F. 2008. The primary target of the killer toxin from Pichia acaciae is tRNA (Gln). Mol Microbiol 69: 681–697. [DOI] [PubMed] [Google Scholar]
- Kuhsel MG, Strickland R, Palmer JD. 1990. An ancient group I intron shared by eubacteria and chloroplasts. Science 250: 1570–1573. [DOI] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johansson MJ, Byström AS. 2005. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 11: 1648–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meineke B, Schwer B, Schaffrath R, Shuman S. 2011. Determinants of eukaryal cell killing by the bacterial ribotoxin PrrC. Nucleic Acids Res 39: 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. 2013. Deregulation of poly(A) polymerase I in Escherichia coli inhibits protein synthesis and leads to cell death. Nucleic Acids Res 41: 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Mejía NE, Florencio-Martínez LE, Figueroa-Angulo EE, Manning-Cela RG, Hernández-Rivas R, Myler PJ, Martínez-Calvillo S. 2009. Gene organization and sequence analyses of transfer RNA genes in Trypanosomatid parasites. BMC Genomics 10: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. 2004. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117: 311–321. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Consaul SA, Nehrke KW, Abelson J. 1992. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J Biol Chem 267: 4577–4582. [PubMed] [Google Scholar]
- Pieńkowska J, Michałowski D, Krzyzosiak WJ, Szweykowska-Kulińska Z. 2002. Pseudouridylation of U35 in the anticodon of Arabidopsis thaliana pre-tRNATyr depends on length rather than structure of an intron. Biochim Biophys Acta 1574: 137–144. [DOI] [PubMed] [Google Scholar]
- Polymenis M, Aramayo R. 2015. Translate to divide: сontrol of the cell cycle by protein synthesis. Microbial Cell 2: 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Will CL, Trowitzsch S, Lührmann R, Söll D, et al. 2011. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331: 760–764. [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Shub DA. 1992. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 357: 173–176. [DOI] [PubMed] [Google Scholar]
- Remus BS, Shuman S. 2013. A kinetic framework for tRNA ligase and enforcement of a 2′-phosphate requirement for ligation highlights the design logic of an RNA repair machine. RNA 19: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MAT, Paris Z, Gaston KW, Fleming IMC, Sample P, Trotta CR, Alfonzo JD. 2013. Unusual noncanonical intron editing is important for tRNA splicing in Trypanosoma brucei. Mol Cell 52: 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya R, Schwer B, Shuman S. 2003. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J Biol Chem 278: 43928–43938. [DOI] [PubMed] [Google Scholar]
- Schneider A, McNally KP, Agabian N. 1993. Splicing and 3′-processing of the tyrosine tRNA of Trypanosoma brucei. J Biol Chem 268: 21868–21874. [PubMed] [Google Scholar]
- Shapiro SZ, Doxsey SJ. 1982. Purification of nuclei from a flagellate protozoan, Trypanosoma brucei. Anal Biochem 127: 112–115. [DOI] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Mark T, Wirtz E, Tschudi C, Ullu E. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu M, Ogawa T, Tanaka W, Takahashi K, Kitamoto HK, Hidaka M, Masaki H. 2013. Evidence for DNA cleavage caused directly by a transfer RNA-targeting toxin. PLoS One 8: e75512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha H, David L, Pascon RC, Clauder-Münster S, Krishnakumar S, Nguyen M, Shi G, Dean J, Davis RW, Oefner PJ, et al. 2008. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180: 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Consaul SA, Phizicky EM. 1997. A conditional lethal yeast phosphotransferase (tpt1) mutant accumulates tRNAs with a 2′-phosphate and an undermodified base at the splice junction. RNA 3: 1388–1400. [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Shuman S. 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286: 7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Meineke B, Shuman S. 2011a. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem 286: 30253–30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Chakravarty AK, Maughan B, Shuman S. 2011b. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J Biol Chem 286: 43134–43143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini GP1, Baldi MI, Gandini-Attardi D, Mattoccia E. 1993. Cleavage site recognition by the tRNA splicing endoribonuclease. Gene 135: 93–97. [DOI] [PubMed] [Google Scholar]
- van Tol H, Beier H. 1988. All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res 16: 1951–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Shuman S. 2005. Structure-function analysis of yeast tRNA ligase. RNA 11: 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem 275: 40174–40179. [DOI] [PubMed] [Google Scholar]
- Wang LK, Lima CD, Shuman S. 2002. Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme. EMBO J 21: 3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland TS, Wickramasekara SM, Guo AY, Selim AL, Winsor TS, Greenleaf AL, Blackwell KL, Olson JA Jr, Marks JR, Bennett CB. 2009. Comparative genome-wide screening identifies a conserved doxorubicin repair network that is diploid specific in Saccharomyces cerevisiae. PLoS One 4: e5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. 2003. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]