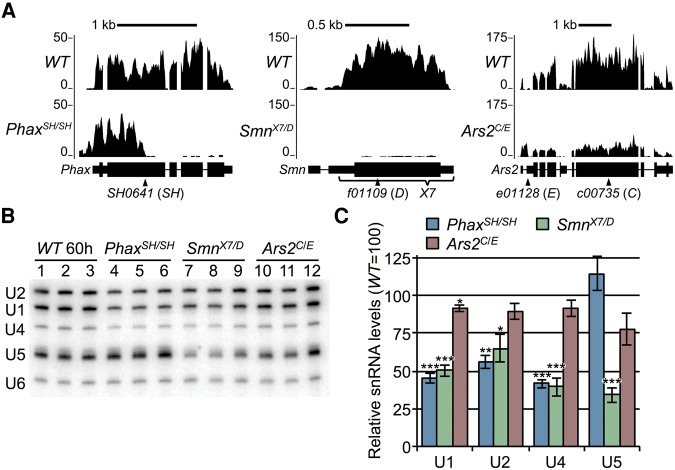
FIGURE 1.
Analysis of three different snRNP biogenesis mutants. (A) Browser shots of larval RNA-seq reads at the three disrupted gene loci. Reads from Oregon-R (WT) versus PhaxSH/SH, SmnX7/D, or Ars2C/E snRNP biogenesis mutants are shown. Mapped read tracks were normalized to the median of the middle two quartiles of the mapped WT sequence read counts. (B) Northern blot of RNA from snRNP biogenesis mutants and WT larvae. RNA was extracted from WT larvae at 60 ± 2 h (60 h) post egg-laying, and mutant RNA was extracted at 74 ± 2 h to account for their delayed development (see text). (C) Quantification of Northern blot in B. Sm-class snRNAs were normalized to the Lsm-class U6 snRNA, and U6-normalized WT snRNA levels were set at 100. Asterisks are P-values from a Student's t-test: (*) P-value ≤0.05, (**) P-value ≤0.01, (***) P-value ≤0.001.