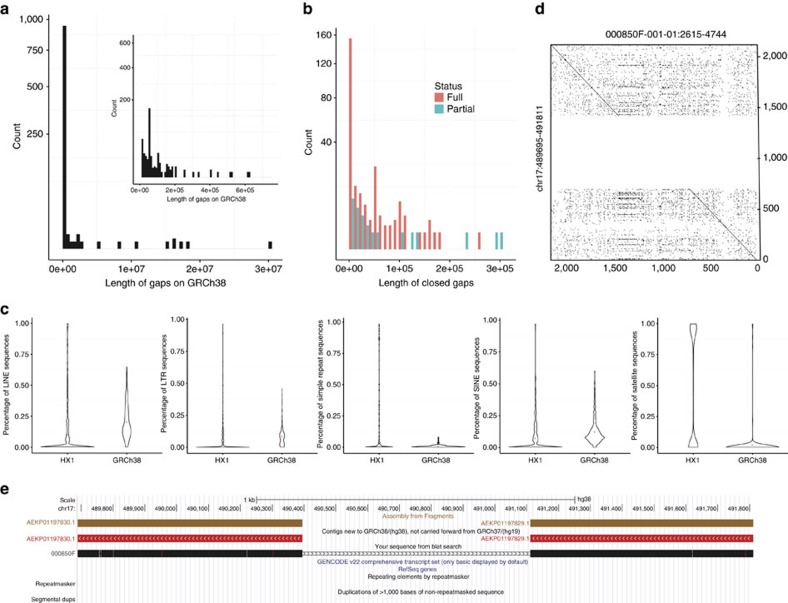
Figure 1. Summary of gap filling in GRCh38.
(a) Length distribution of all gaps (stretches of ‘N' in genome sequence) in GRCh38. (b) Length distribution of all gaps that can be fully or partially closed. (c) Violin plots showing the distribution of LINE, SINE, LTR, simple repeat and satellite in closed gaps and in GRCh38. (d) A dotplot showing how a gap on 17p13.3 is closed by a contig in HX1. The plot shows comparison of two sequences and each dot indicates a region of close similarity between them. (e) Genome browser screenshot of the gap region that was closed. The gap is flanked by two contigs that are new in GRCh38 (not carried forward from GRCh37), yet an HX1 associated contig (000850F-001-01) can completely align to flanking regions, therefore filling this assembly gap and revising its length from 718 to 731 bp.