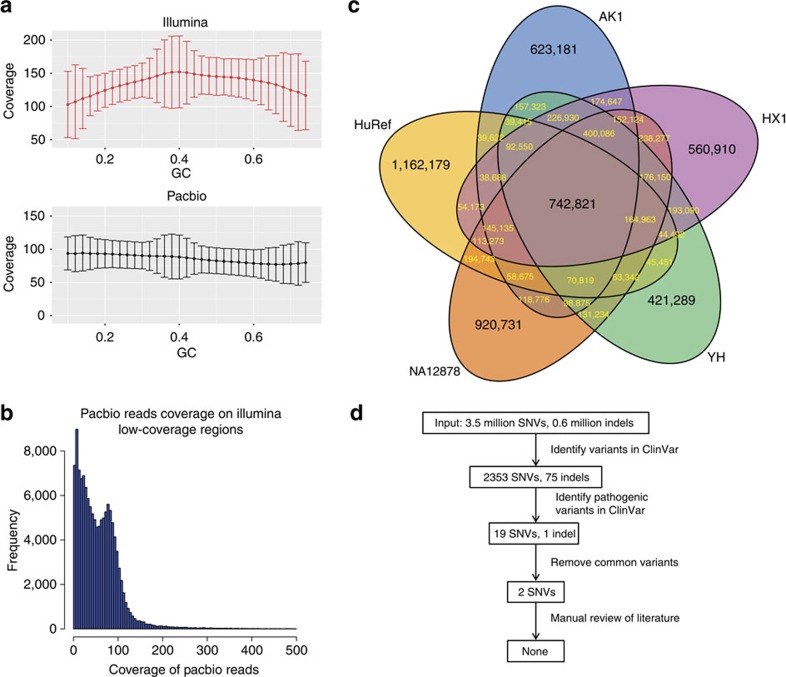
Figure 4. Functional annotation and analysis of the genomic variants in HX1.
(a) Average coverage versus GC contents for 100-bp windows in Illumina data and PacBio data, respectively. The mean and s.d. values are shown. (b) Distribution of PacBio coverage for regions that have ≤5 × coverage in Illumina data. (c) Shared SNVs discovered in HX1, AK1, HuRef, NA12878 and YH. (d) Variant reduction pipeline to identify pathogenic variant; although 20 were annotated as ‘pathogenic' in ClinVar, careful analysis failed to support any one.