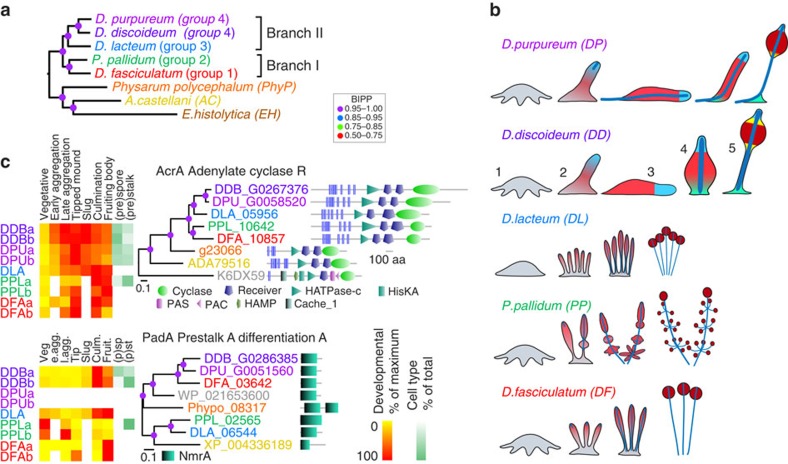
Figure 1. Dictyostelid phylogeny with life cycles and selected annotated gene trees.
(a) Phylogenetic tree of dictyostelid and unicellular amoebozoan species with sequenced genomes, as inferred from 30 concatenated proteins by Bayesian inference3. (b) Schematic of life cycle complexity of the Dictyostelid test species. DF, PP and DL form multiple fruiting bodies directly from the aggregate. All cells first differentiate into prespore cells and then form the stalk by dedifferentiation of prespore cells at the tip. DD and DP form single fruiting bodies from aggregates and display an intermediate migratory ‘slug' in which cells pre-differentiate into prestalk and prespore cells. During fruiting body formation, two more cell types emerge that support the stalk and spore mass3,4. 1: aggregate, 2: early sorogen (slug), 3: migrating slug, 4: mid-culminant, 5: fruiting body. Light red: prespore; dark red: spore; light blue: prestalk; dark blue: stalk; green: basal disc or supporter; yellow: upper and lower cup. (c) Annotated gene trees. Examples of trees, based on the amino-acid sequence of homologues of DD DEG, annotated with protein functional domains and developmental expression profiles and inferred by Bayesian inference (see Methods for procedures). The AcrA tree follows the amoebozoan tree topology, identifying all amoebozoan homologs as orthologues. The PadA tree does not, identifying PPL_02565 and DLA_06544 as paralogues of DD padA.