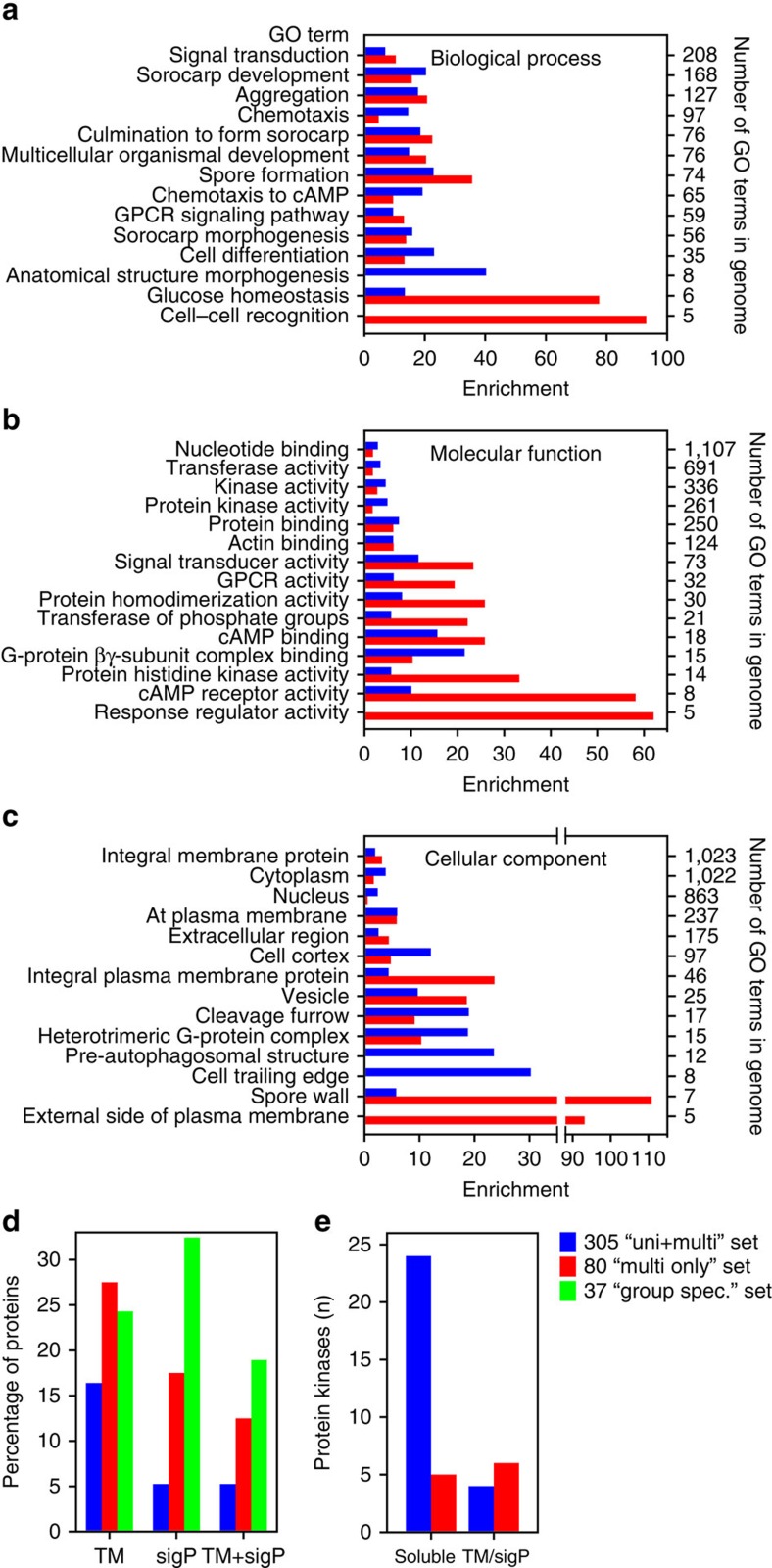
Figure 4. GO enrichment, signal peptides (SigP) and transmembrane (TM) domains.
(a-c) Enrichment of GO terms in the categories ‘biological process' (a), ‘molecular function (b) and ‘cellular component' (c). The sets of 305 and 80 DEGs, which are respectively with and without homologues in unicellular amoebozoa, were analysed for GO term enrichment (Supplementary Data 3). The 10 terms with lowest P values in each set are compared with the same terms in the other set. The enrichment factor (frequency in set/frequency in genome) for each term is shown. Note that for GO terms that are very abundant in the genome (numbers on right axis) low-enrichment factors are still associated with very low P values. (d) SigP and TM domains. Proteins in the 305 and 80 sets, as well as 37 proteins with limited conservation within Dictyostelia (green) were analysed with Phobius63 for transmembrane domains and signal peptides (Supplementary Data 4). Percentages of proteins with either SigP or TM domains, or with both are presented. (e) Protein kinase localization. All protein kinases in the 305 and 80 sets (S/T/Y, histidine and alpha kinases) were analysed with Phobius (Supplementary Data 4) and numbers of proteins in each set without SigP or TM domains (soluble) and with either TM or SigP domains are presented.