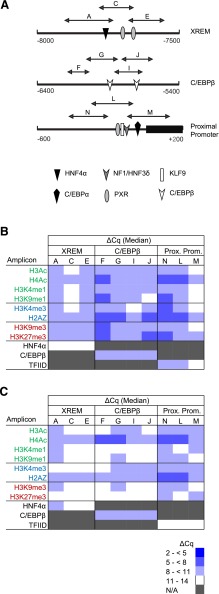
Fig. 5.
CYP3A7 promoter occupancy by modified and variant histones or transcription factors. (A) Shown are the three known CYP3A7 regulatory domains probed in the current study, all known transcription factors binding within each domain, and the qPCR amplicons used to evaluate ChIP enrichment. ChIP was performed with antibodies for modified and variant histones, as well as a single transcription factor known to bind within each domain, followed by qPCR to quantify occupancy. The data for pooled fetal (N = 14) (B) and postnatal (N = 10) (C) hepatic chromatin are shown as blue-tinted heat maps representing a range of median ΔCq values from a minimum of three experiments for each amplicon within the indicated CYP3A7 region. N/A, not assayed. Modified and variant histones are grouped into those associated with active (green font), poised (blue font), or inactive transcription (red font). The relevant occupancy by select transcription factors (black font) is also shown. HNF4, hepatocyte nuclear factor 4; KLF9, Kruppel-like factor 9; NF1/HNF3, nuclear factor 1/hepatocyte nuclear factor 3; PXR, pregnane X receptor.