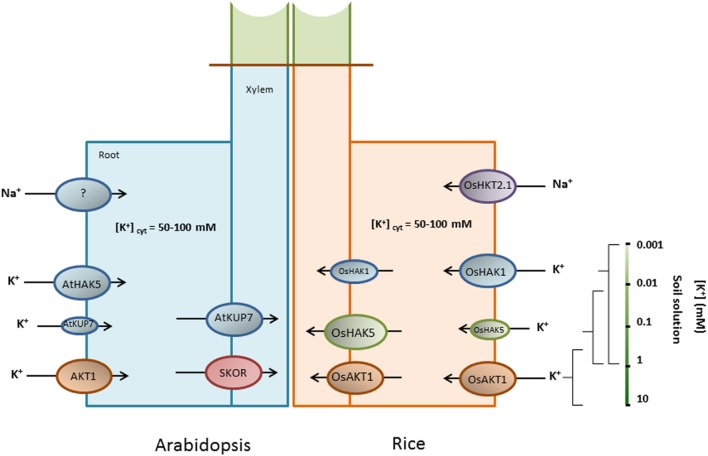
FIGURE 1.
Schematic comparison of the systems involved in K+ and Na+ movements in Arabidopsis and rice roots. The availability of knock-out mutants in Arabidopsis and rice plants for specific transport systems has allowed for elucidating their roles in K+ transport. The figure shows the predicted function of each system for which knock-out mutants have been studied. AtHAK5 and AKT1 are the main systems for K+ uptake in Arabidopsis plants. In addition, a member of the KT/HAK/KUP family, AtKUP7, seems to also be involved in K+ uptake. K+ release into the xylem is mainly mediated by SKOR and also partially by AtKUP7. In rice, AtHAK5 and AKT1 functions are fulfilled by the rice homologs OsHAK1 and OsAKT1. An additional system, OsHAK5, partially contributes to high-affinity K+ uptake, but at higher concentrations than OsHAK1. These three rice systems may directly or indirectly facilitate K+ release into the xylem, with the contribution by OsHAK5 to K+ release into the xylem being more relevant. It is not clear if such contribution is a direct (by mediating K+ efflux into xylem vessels) or indirect (by favoring K+ accumulation in endodermal cells) outcome of the aforementioned transporters. Regarding Na+ uptake, the genetic identity of Na+ uptake systems in Arabidopsis remains to be elucidated. GLRs, CNGCs or other non-selective cation channels could be involved. In rice, OsHKT2;1 has been shown to mediate Na+ uptake during K+ deficiency. In the presence of K+ or high external Na+ concentrations other unknown systems should take part in Na+ uptake.