Abstract
There is no information regarding the role of microRNAs in the development of the external ear in mammals. The purpose of this study was to determine the stage-specific expression of microRNA during external ear development in mice under normal conditions. GeneChip miRNA 3.0 arrays by Affymetrix were used to obtain miRNA expression profiles from mice fetal pinnae and back skin tissues at 13.5 days-post-coitum (dpc) and 14.5 dpc. Biological triplicates for each tissue were analyzed; one litter represents one biological replica, each litter had 16 fetuses on average. The results were analyzed with Affymetrix's Transcriptome Analysis Console software to identify differentially expressed miRNAs. The inquiry showed significant differential expression of 25 miRNAs at 13.5 dpc and 31 at 14.5 dpc, some of these miRNAs were predicted to target genes implicated in external ear development. One example is mmu-miR-10a whose low expression in pinnae is known to impact ear development by modulating Hoxa1 mRNA levels Garzon et al. (2006), Gavalas et al. (1998) [1], [2]. Other findings like the upregulation of mmu-miR-200c and mmu-miR-205 in the pinnae tissues of healthy mice are in agreement with what has been reported in human patients with microtia, in which down regulation of both miRNAs has been found Li et al. (2013) [3].
This study uncovered a spatiotemporal pattern of miRNA expression in the external ear, which results from continuous transcriptional changes during normal development of body structures.
All microarray data are available at the Gene Expression Omnibus (GEO) at NCBI under accession number GSE64945.
Keywords: Microarray, miRNAs, Mouse, Ear development
| Specifications | |
| Organism/cell line/tissue | Mus musculus CD1 strain/fetuses/pinnae and back skin |
| Sex | Male and female |
| Sequencer or array type | Affymetrix GeneChip miRNA 3.0 array |
| Data format | Raw CEL and CHP files |
| Experimental factors | Developmental stages: 13.5 and 14.5 dpc |
| Experimental features | Total RNA from external ear tissue, the pinnae (cartilage and skin), and back skin tissues of each developmental stage was extracted and subjected to microarray analysis. The raw data were analyzed with Expression Console and Transcriptome Analysis Console by Affymetrix. |
| Consent | Approved by the Institutional committee of Care and Use of Laboratory Animals, Instituto Nacional de Pediatría, México. |
| Sample source location | Not applicable |
1. Direct link to deposited data
The data are available at GEO public repository:
2. Experimental design, materials and methods
2.1. Mice
In this study we worked with CD1 mice strain. The mice habitat conditions were controlled (12 h:12 h light/dark cycle, temperature 19.5–22.5 °C, relative humidity 45–55%) and the mice were provided with water and food (Teklab Global Rodent Diet, Harlan) ad libitum. All management and experimental procedures were in accordance with the Mexican regulation norm NOM-062-ZOO-1999 and the Guide for Care and Use of Laboratory Animals, Institute for Laboratory Animal Research of the National Academy of Sciences, USA. The Institutional Animal Care and Use of Laboratory Animals and Research Committees from the Instituto Nacional de Pediatría, approved this study.
Scheduled mating was carried out in order to obtain fetuses at specific developmental stages. Pregnant females were sacrificed by cervical dislocation at 13.5 dpc or 14.5 dpc. The gestational age of the fetus was determined by the day of appearance of the vaginal plug (0.5 dpc) and confirmed by the following morphological criteria:
| Gestational age | Somite pair count | Limbs | Fur |
|---|---|---|---|
| 13.5 | 52–55 | Distal separation of the fingers | Absence of hair follicles |
| 14.5 | 56–60 | Proximal separation of the fingers | Sparse facial hair |
2.2. Tissues
Biological triplicates for each tissue and each developmental stage were analyzed. One litter represents one biological replica and each litter had an average of 16 fetuses (32 pinnae). External ear tissue, the pinnae (cartilage and skin), and back skin tissues were dissected under stereomicroscope in iced cooled nuclease free PBS (Gibco, Waltham, MA USA) and were kept in RNAlater (Ambion, Waltham, MA USA) at -80 °C until further processing. All tissues belonging to a specific body region from a particular developmental stage were pooled in a single sample per biological replica; this procedure was very successful to obtain enough tissue from this anatomically small region.
2.3. microRNA array analysis
Total RNA including short RNAs (< 200 bp) was extracted with an RNA Mini elute kit (Qiagen, Venlo, Netherlands) and quantitated with a NanodropTM spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). RNA quality was monitored by agarose gel electrophoresis.
miRNA profiling in the extracted RNA was performed by using the Affymetrix GeneChip miRNA Arrays 3.0 (Affymetrix, Santa Clara, CA, USA). In this version of the array the manufacturer includes 179,217 probes that represent 19,913 mature miRNA contained in miRBase V.17 (www.mirbase.org) with probes for 153 different organisms, including 1111 probes for Mus musculus miRNAs and 855 pre-miRNAs. We performed the arrays according to Affymetrix instructions. Briefly, 500 ng of total RNA, were labeled with the FlashTag ® Biotin HSR kit (Genisphere/Affymetrix, Santa Clara, CA, USA). Correct labeling was confirmed by Enzyme Linked Oligosorbent Assay (ELOSA) QC Assay (Thermo Fisher Scientific Inc., Waltham, MA, USA). Samples were denaturalized at 99 °C for 5 min followed by 45 °C for another 5 min, injected into the array chips and allowed hybridization for 17 h at 48 °C in an Affymetrix Hybridization Oven 645 in constant movement at 60 rpm. Arrays were then stained and washed in the Affymetrix GeneChip Fluidic Station 450 and scanned using an Affymetrix GeneChip Scanner 3000 7G (Affymetrix, Santa Clara, CA, USA).
The miRNA QC tool was used to assess the quality of the array data, afterwards data were normalized by global normalization with Robust Multichip Average (RMA) and Detection Above Background (DABG) using of the Expression Console software (Affymetrix, Santa Clara, CA, USA) (Fig. 1, Fig. 2). The identification of differentially expressed miRNAs at each stage, was achieved by using linear regression and hierarchical clustering analysis with the Transcriptome Analysis Console software from Affymetrix. The filter settings were the following: “Transcription Cluster ID”: contains mmu-miR does not contain hp.-mmu-miR, “Fold change”: + 3 and − 3, “False discovered rate (FDR)”: 1.0. “Adjusted P-value threshold”: 0.05. Expression levels of the miRNAs were compared between ear and back tissues (Fig. 3, Fig. 4, Fig. 5, Fig. 6) (Table 1, Table 2).
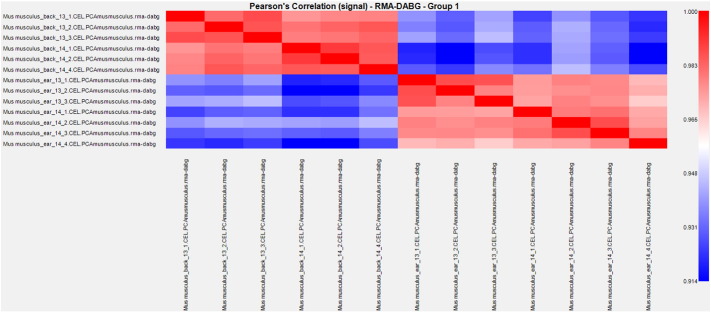
Fig. 1.
Pearson's Correlation of the signal obtained from Affymetrix miRNA Arrays 3.0 GeneChips hybridized.
Fig. 2.
Loading plot showing the components from Principal Component Analysis (PCA) of all probe sets from Affymetrix GeneChip miRNA Arrays 3.0 hybridized. We clearly observed four groups that can be arranged by developmental stage (red cubes 13.5 dpc, red cubes with black edges 14.5 dpc) or by tissue (right side pinnae, left side back skin).
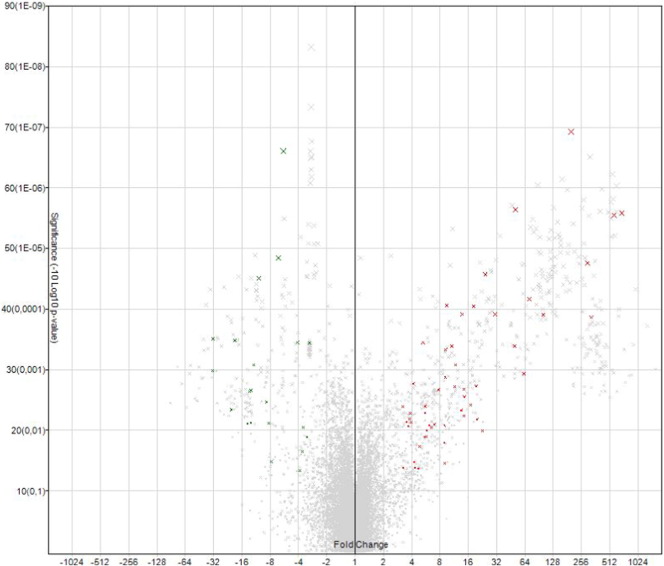
Fig. 3.
Volcano plot from GeneChip miRNA Arrays 3.0 analysis at 13.5 dpc, expression levels comparing ear (pinnae) and back tissues. Fold change: + 3 and − 3, Adjusted P-value threshold 0.05. Significant spots in green and red, green X: low expression levels in back skin, red X: high expression levels in pinnae.
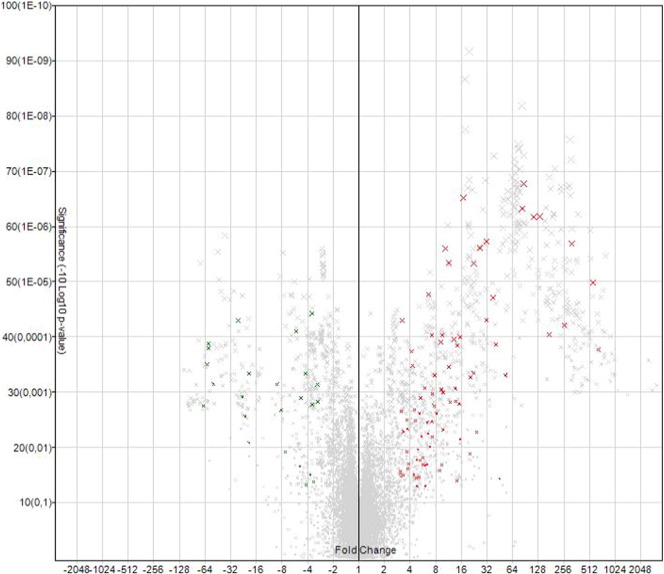
Fig. 4.
Volcano plot from GeneChip miRNA Arrays 3.0 analysis at 14.5 dpc, expression levels comparing ear (pinnae) and back tissues. Fold change: + 3 and − 3, Adjusted P-value threshold 0.05. Significant spots in green and red, green X: low expression levels in back skin, red X: high expression levels in pinnae.
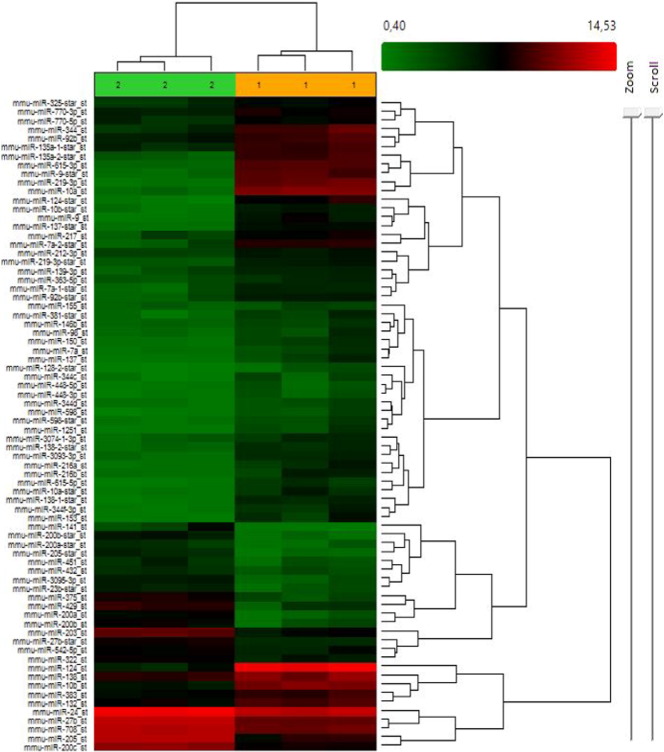
Fig. 5.
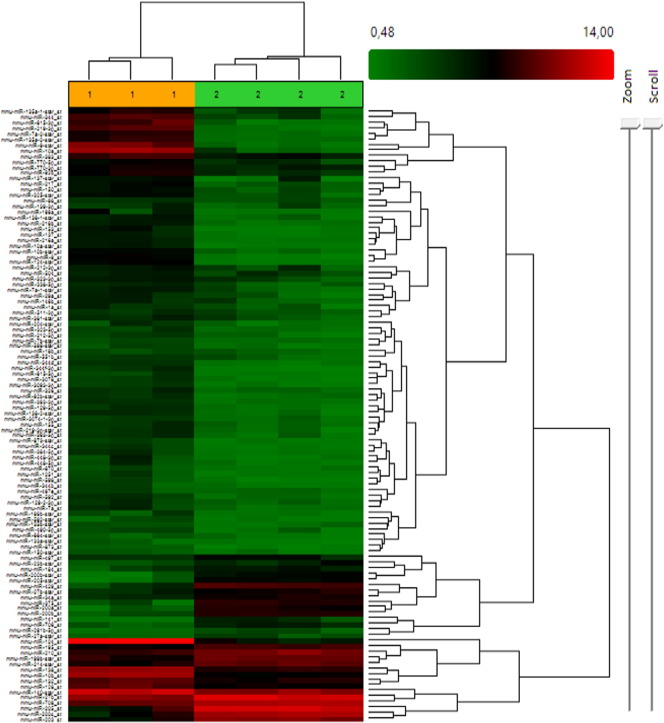
Heat map from GeneChip miRNA Arrays 3.0 analysis at 13.5 dpc, expression levels comparing ear (pinnae)(2) and back tissues(1). Fold change: + 3 and − 3, Adjusted P-value threshold 0.05. Green low expression levels in back skin, red high expression levels in pinnae.
Fig. 6.
Heat map from GeneChip miRNA Arrays 3.0 analysis at 14.5 dpc, expression levels comparing ear (pinnae)(2) and back tissues (1). Fold change: + 3 and − 3, Adjusted P-value threshold 0.05. Green low expression levels in back skin, red high expression levels in pinnae.
Table 1.
Differentially expressed microRNA in ear (pinnae) versus back skin at 13.5 dpc.
| microRNA ID | Ear 13.5 dpc | Back 13.5 dpc | Fold change |
|---|---|---|---|
| mmu-miR-200a_st | 7.14 | 2.1 | 32.85 |
| mmu-miR-200b-star_st | 6.23 | 1.21 | 32.44 |
| mmu-miR-200b_st | 7.41 | 3.03 | 20.76 |
| mmu-miR-375_st | 8.09 | 3.84 | 19 |
| mmu-miR-200a-star_st | 5.13 | 1.32 | 14.01 |
| mmu-miR-3095-3p_st | 6.07 | 2.35 | 13.25 |
| mmu-miR-429_st | 8.54 | 4.85 | 12.96 |
| mmu-miR-205_st | 12.44 | 8.75 | 12.85 |
| mmu-miR-205-star_st | 5.31 | 1.72 | 12.05 |
| mmu-miR-200c_st | 11.28 | 7.86 | 10.67 |
| mmu-miR-203_st | 10.07 | 6.94 | 8.75 |
| mmu-miR-23b-star_st | 5.82 | 2.77 | 8.3 |
| mmu-miR-141_st | 3.93 | 0.97 | 7.79 |
| mmu-miR-27b-star_st | 7.92 | 5.22 | 6.53 |
| mmu-miR-27b_st | 12.69 | 10.15 | 5.82 |
| mmu-miR-708_st | 12.39 | 10.36 | 4.1 |
| mmu-miR-542-5p_st | 7.45 | 5.5 | 3.88 |
| mmu-miR-451_st | 4.68 | 2.82 | 3.63 |
| mmu-miR-432_st | 4.98 | 3.14 | 3.56 |
| mmu-miR-322_st | 7.65 | 5.95 | 3.25 |
| mmu-miR-24_st | 14.13 | 12.52 | 3.05 |
| mmu-miR-383_st | 7.14 | 8.82 | − 3.2 |
| mmu-miR-155_st | 2.1 | 3.8 | − 3.26 |
| mmu-miR-344d_st | 1.39 | 3.21 | − 3.53 |
| mmu-miR-598_st | 0.8 | 2.67 | − 3.65 |
| mmu-miR-598-star_st | 1.29 | 3.21 | − 3.78 |
| mmu-miR-132_st | 7.48 | 9.44 | − 3.89 |
| mmu-miR-146b_st | 1.91 | 3.9 | − 3.95 |
| mmu-miR-770-3p_st | 5.78 | 7.85 | − 4.2 |
| mmu-miR-448-5p_st | 0.86 | 2.95 | − 4.25 |
| mmu-miR-98_st | 1.58 | 3.7 | − 4.34 |
| mmu-miR-128-2-star_st | 0.57 | 2.81 | − 4.73 |
| mmu-miR-448-3p_st | 0.89 | 3.16 | − 4.83 |
| mmu-miR-212-3p_st | 3.91 | 6.3 | − 5.27 |
| mmu-miR-139-3p_st | 3.12 | 5.57 | − 5.47 |
| mmu-miR-363-5p_st | 2.87 | 5.34 | − 5.54 |
| mmu-miR-138_st | 8.61 | 11.1 | − 5.58 |
| mmu-miR-7a_st | 1.26 | 3.78 | − 5.72 |
| mmu-miR-381-star_st | 1.76 | 4.29 | − 5.79 |
| mmu-miR-770-5p_st | 4.71 | 7.33 | − 6.15 |
| mmu-miR-1251_st | 0.86 | 3.56 | − 6.47 |
| mmu-miR-325-star_st | 4.31 | 7.11 | − 6.99 |
| mmu-let-7b_st | 11.04 | 13.96 | − 7.6 |
| mmu-miR-150_st | 1.33 | 4.28 | − 7.71 |
| mmu-miR-137_st | 1.07 | 4.22 | − 8.88 |
| mmu-miR-3074-1-3p_st | 2.34 | 5.5 | − 8.91 |
| mmu-miR-344c_st | 0.72 | 3.88 | − 8.95 |
| mmu-miR-92b_st | 5.93 | 9.12 | − 9.09 |
| mmu-miR-138-2-star_st | 1.93 | 5.13 | − 9.19 |
| mmu-miR-219-3p-star_st | 2.78 | 6.02 | − 9.41 |
| mmu-miR-615-5p_st | 0.81 | 4.22 | − 10.65 |
| mmu-miR-344f-3p_st | 1.03 | 4.55 | − 11.47 |
| mmu-miR-135a-1-star_st | 5.4 | 8.95 | − 11.73 |
| mmu-miR-216a_st | 1.32 | 5.07 | − 13.5 |
| mmu-miR-3093-3p_st | 1.43 | 5.2 | − 13.68 |
| mmu-miR-10a-star_st | 1.12 | 4.97 | − 14.42 |
| mmu-miR-7a-1-star_st | 1.67 | 5.53 | − 14.47 |
| mmu-miR-217_st | 3.47 | 7.33 | − 14.51 |
| mmu-miR-92b-star_st | 1.99 | 6.08 | − 17.04 |
| mmu-miR-10b_st | 6.63 | 10.81 | − 18.21 |
| mmu-miR-344_st | 4.8 | 9.08 | − 19.38 |
| mmu-miR-153_st | 0.86 | 5.18 | − 19.92 |
| mmu-miR-216b_st | 1.42 | 5.91 | − 22.6 |
| mmu-miR-138-1-star_st | 0.88 | 5.47 | − 24.21 |
| mmu-miR-10b-star_st | 0.93 | 5.88 | − 30.85 |
| mmu-miR-9_st | 0.87 | 6.51 | − 49.6 |
| mmu-miR-137-star_st | 1.27 | 6.93 | − 50.77 |
| mmu-miR-7a-2-star_st | 2.48 | 8.45 | − 62.68 |
| mmu-miR-135a-2-star_st | 2.68 | 8.84 | − 71.33 |
| mmu-miR-124-star_st | 0.81 | 7.46 | − 99.99 |
| mmu-miR-615-3p_st | 1.96 | 9.58 | − 197.55 |
| mmu-miR-9-star_st | 1.51 | 9.72 | − 296.06 |
| mmu-miR-124_st | 5.85 | 14.2 | − 325.39 |
| mmu-miR-219-3p_st | 1.12 | 10.27 | − 565.86 |
| mmu-miR-10a_st | 1.57 | 10.99 | − 684.75 |
Table 2.
Differentially expressed microRNA in ear (pinnae) versus back skin at 14.5 dpc.
| microRNA ID | Ear 14.5 dpc | Back 14.5 dpc | Fold change |
|---|---|---|---|
| mmu-miR-205_st | 12.87 | 6.8 | 67.04 |
| mmu-miR-200b-star_st | 6.78 | 0.86 | 60.56 |
| mmu-miR-205-star_st | 6.94 | 1.08 | 58.14 |
| mmu-miR-200a_st | 7.93 | 2.07 | 57.91 |
| mmu-miR-375_st | 8.27 | 2.6 | 50.82 |
| mmu-miR-200b_st | 8.2 | 3.49 | 26.15 |
| mmu-miR-429_st | 9.1 | 4.54 | 23.51 |
| mmu-miR-200c_st | 11.6 | 7.16 | 21.63 |
| mmu-miR-141_st | 5.43 | 1.14 | 19.67 |
| mmu-miR-203_st | 10.84 | 6.56 | 19.51 |
| mmu-miR-706_st | 5.61 | 2.41 | 9.15 |
| mmu-miR-27b-star_st | 7.65 | 4.63 | 8.14 |
| mmu-miR-23b-star_st | 5.39 | 2.52 | 7.31 |
| mmu-miR-34a_st | 7.75 | 5.3 | 5.48 |
| mmu-miR-184_st | 5.98 | 3.69 | 4.89 |
| mmu-miR-199b-star_st | 9.91 | 7.65 | 4.79 |
| mmu-miR-27b_st | 12.88 | 10.81 | 4.19 |
| mmu-miR-497_st | 6.72 | 4.66 | 4.17 |
| mmu-miR-291b-5p_st | 3.56 | 1.67 | 3.72 |
| mmu-miR-708_st | 11.69 | 9.86 | 3.56 |
| mmu-miR-210_st | 10.18 | 8.37 | 3.52 |
| mmu-miR-27a-star_st | 3.66 | 1.9 | 3.4 |
| mmu-miR-195_st | 9.07 | 7.45 | 3.07 |
| mmu-miR-214-star_st | 9.69 | 8.08 | 3.05 |
| mmu-miR-18b_st | 2.03 | 3.64 | − 3.04 |
| mmu-miR-664-star_st | 1.04 | 2.66 | − 3.06 |
| mmu-miR-132_st | 7.92 | 9.58 | − 3.15 |
| mmu-miR-133a-star_st | 0.82 | 2.47 | − 3.15 |
| mmu-miR-873_st | 0.71 | 2.39 | − 3.22 |
| mmu-let-7b_st | 12.01 | 13.74 | − 3.3 |
| mmu-miR-150-star_st | 1.03 | 2.76 | − 3.32 |
| mmu-miR-511-3p_st | 2.84 | 4.58 | − 3.33 |
| mmu-miR-670_st | 1.21 | 3.07 | − 3.63 |
| mmu-miR-135b-star_st | 1.18 | 3.06 | − 3.68 |
| mmu-miR-98_st | 2.78 | 4.68 | − 3.73 |
| mmu-miR-1a_st | 2.63 | 4.54 | − 3.78 |
| mmu-miR-448-3p_st | 0.93 | 2.88 | − 3.84 |
| mmu-miR-7b-star_st | 1.6 | 3.67 | − 4.2 |
| mmu-miR-128_st | 7.37 | 9.47 | − 4.28 |
| mmu-miR-504_st | 3.82 | 5.97 | − 4.43 |
| mmu-miR-323-5p_st | 1.67 | 3.84 | − 4.5 |
| mmu-miR-140-star_st | 10.51 | 12.73 | − 4.64 |
| mmu-miR-381-star_st | 2.87 | 5.12 | − 4.75 |
| mmu-miR-196b-star_st | 1.22 | 3.49 | − 4.8 |
| mmu-miR-467e_st | 1.89 | 4.16 | − 4.8 |
| mmu-miR-139-3p_st | 1.78 | 4.07 | − 4.9 |
| mmu-miR-129-2-3p_st | 2.04 | 4.36 | − 4.97 |
| mmu-miR-323-3p_st | 3.61 | 5.96 | − 5.11 |
| mmu-miR-770-3p_st | 5.39 | 7.76 | − 5.17 |
| mmu-miR-551b_st | 2.02 | 4.42 | − 5.25 |
| mmu-miR-155_st | 1.86 | 4.27 | − 5.29 |
| mmu-miR-1251_st | 1.07 | 3.51 | − 5.42 |
| mmu-miR-448-5p_st | 0.92 | 3.42 | − 5.66 |
| mmu-miR-770-5p_st | 4.97 | 7.48 | − 5.71 |
| mmu-miR-615-5p_st | 0.9 | 3.48 | − 5.94 |
| mmu-miR-204-star_st | 1.52 | 4.1 | − 6.02 |
| mmu-miR-212-3p_st | 3.46 | 6.06 | − 6.06 |
| mmu-miR-592-star_st | 0.84 | 3.51 | − 6.35 |
| mmu-miR-598-star_st | 1.21 | 3.88 | − 6.36 |
| mmu-miR-212-5p_st | 1.52 | 4.22 | − 6.5 |
| mmu-miR-138_st | 8.31 | 11.04 | − 6.63 |
| mmu-miR-490-5p_st | 1.01 | 3.78 | − 6.85 |
| mmu-miR-873-star_st | 0.94 | 3.79 | − 7.21 |
| mmu-miR-598_st | 0.75 | 3.61 | − 7.22 |
| mmu-miR-3074-1-3p_st | 1.41 | 4.27 | − 7.27 |
| mmu-miR-338-5p_st | 2.41 | 5.28 | − 7.33 |
| mmu-miR-383_st | 6.16 | 9.09 | − 7.65 |
| mmu-miR-326_st | 1.87 | 4.83 | − 7.8 |
| mmu-miR-219-3p-star_st | 1.62 | 4.65 | − 8.18 |
| mmu-miR-344b_st | 0.92 | 4.09 | − 8.99 |
| mmu-miR-92b-star_st | 1.45 | 4.64 | − 9.16 |
| mmu-miR-3078_st | 0.85 | 4.05 | − 9.18 |
| mmu-miR-92b_st | 4.88 | 8.09 | − 9.28 |
| mmu-miR-29a_st | 2.22 | 5.45 | − 9.37 |
| mmu-miR-3093-3p_st | 0.83 | 4.08 | − 9.5 |
| mmu-miR-7a_st | 1.53 | 4.81 | − 9.67 |
| mmu-miR-592_st | 1.59 | 4.88 | − 9.77 |
| mmu-miR-363-3p_st | 1.37 | 4.74 | − 10.34 |
| mmu-miR-344d_st | 1.16 | 4.68 | − 11.45 |
| mmu-miR-344c_st | 0.88 | 4.4 | − 11.5 |
| mmu-miR-363-5p_st | 1.18 | 4.74 | − 11.81 |
| mmu-miR-129-5p_st | 1.31 | 5.04 | − 13.21 |
| mmu-miR-384-5p_st | 1.11 | 4.86 | − 13.49 |
| mmu-miR-150_st | 2.64 | 6.44 | − 13.86 |
| mmu-miR-146b_st | 1.87 | 5.69 | − 14.16 |
| mmu-miR-138-2-star_st | 1.3 | 5.14 | − 14.35 |
| mmu-miR-325-star_st | 2.86 | 6.77 | − 15.09 |
| mmu-miR-7a-1-star_st | 1.9 | 5.84 | − 15.38 |
| mmu-miR-344f-3p_st | 0.65 | 4.61 | − 15.65 |
| mmu-miR-10b_st | 7.3 | 11.38 | − 16.94 |
| mmu-miR-196a_st | 1.16 | 5.51 | − 20.32 |
| mmu-miR-138-1-star_st | 1.49 | 5.85 | − 20.52 |
| mmu-miR-217_st | 2.42 | 6.9 | − 22.28 |
| mmu-miR-216b_st | 1.48 | 5.96 | − 22.35 |
| mmu-miR-135a-1-star_st | 3.47 | 8.08 | − 24.38 |
| mmu-miR-216a_st | 1.24 | 5.96 | − 26.29 |
| mmu-miR-137_st | 0.99 | 5.97 | − 31.56 |
| mmu-miR-10a-star_st | 0.87 | 5.86 | − 31.75 |
| mmu-miR-10b-star_st | 1.68 | 6.91 | − 37.5 |
| mmu-miR-153_st | 0.69 | 6.03 | − 40.5 |
| mmu-miR-137-star_st | 0.84 | 6.33 | − 44.98 |
| mmu-miR-344_st | 3.49 | 9.2 | − 52.37 |
| mmu-miR-9_st | 0.79 | 7.16 | − 82.76 |
| mmu-miR-124-star_st | 0.92 | 7.36 | − 86.83 |
| mmu-miR-7a-2-star_st | 1.46 | 8.27 | − 112.48 |
| mmu-miR-135a-2-star_st | 1.56 | 8.61 | − 132.71 |
| mmu-miR-615-3p_st | 1.13 | 8.55 | − 171.31 |
| mmu-miR-124_st | 5.98 | 13.98 | − 256.55 |
| mmu-miR-219-3p_st | 1.27 | 9.57 | − 316.41 |
| mmu-miR-9-star_st | 1.4 | 10.54 | − 562.1 |
| mmu-miR-10a_st | 1.83 | 11.18 | − 651.42 |
Putative mRNA targets for differentially expressed miRNAs were identified using the bioinformatic predicting tools TargetScanMouse Release 6.2 (www.targetscan.org) miRNA body Map (www.mirnabodymap.org), miRBase (www.mirbase.org) and DIANA Tools (Tarbase, microT v4 and microT-CDS), (http://diana.imis.athena-innovation.gr/DianaTools) (Table 3).
Table 3.
Putative mRNA targets for differentially expressed miRNAs.
| Bioinformatic predicting tools/mmu-miR differentially expressed |
||||
|---|---|---|---|---|
| Target Gene | miRNA body Map v1.1 | microT v4 | microT-CDS v5 | Tarbase |
| Bmp5 | mmu-miR-203 | – | – | – |
| Cyp26b1 | mmu-miR-24, 200a, 205, 9, 217 | – | mmu-miR-195, 200ª | – |
| Dlx5 | mmu-miR-124, 203, 200a | – | mmu-miR-124 | mmu-miR-200a |
| Dlx6 | mmu-miR-205 | – | mmu-miR-7a-2-star | – |
| Edn1 | mmu-miR-203 | – | mm-miR-344d, 135a-2-star | – |
| Ednra | mmu-miR-203, 27b, 324-3p, 497, 195, 124 | – | mmu-miR-27b | mmu-miR-324-3p |
| Eya1 | mmu-miR-23b, 27b, 200a, 203, 205, 497, 195, 124, 217 | mmu-miR-27b | mmu-miR-124, 10a-star, 195, 497, 27b | – |
| Fgf8 | mmu-miR-195 | – | – | – |
| Fgf10 | mmu-miR-9, 200c, 200b, 203, 375, 429, 137, 217, 344 | – | mmu-miR-137, 124, 9 | – |
| Frem2 | mmu-miR-9, 200b, 24, 205, 27b, 200c, 429, 216a, | – | mmu-miR-9, 429, 200c, 200b, 24 | – |
| Gsc | mmu-miR-200c | – | – | – |
| Hoxa1 | mmu-miR-23b, 28, 203, 9, 10a, 216a | – | mmu-miR-23b | – |
| Hoxa2 | mmu-miR-23b | – | – | – |
| Irf6 | mmu-miR-24, 27b, 200c, 200b, 324-3p, 429, 216a, | – | – | – |
| Pax8 | mmu-miR-203, 9 | – | – | – |
| Prrx1 | mmu-miR-203, 205, 375, 9, 10a, 124 | – | mmu-miR-3093-3p, 124, 9, 195, 497, 375 | – |
| Prrx2 | mmu-miR-124, 216a | – | – | |
| Prkra | mmu-miR-10a, 10b | – | mmu-miR-10b, 27b-star | – |
| Sall1 | mmu-miR-195, 497, 28, 205 | – | mmu-miR-195, 497 | – |
| Six1 | mmu-miR-27b, 200a, 200b, 200c, 205, 375, 429, 10a, 217 | – | mmu-miR-217, 7a-2-star, 429, 375, 200a | – |
| Six4 | mmu-miR-23b, 24, 27b, 200a, 200b, 429, 497, 195, 9, 10a, 124, 216a, 217 | mmu-miR-124, 9 | mm-miR-344d, 124, 9, 205-star, 27b, 23b | – |
| Tcfap2a | mmu-miR-195, 200a, 200b, 200c, 375, 429, 497, 137 | – | mmu-miR-137, 124-star, 497, 429, 200c, 200b | – |
| Wnt5a | mmu-miR-205, 375 | – | mmu-miR-205-star, 200a | – |
| Chuk | mmu-miR-23b, 124, 200a, | – | mmu-miR-124, 23b | – |
| Hoxb1 | – | – | mmu-miR-124, 375 | – |
3. Discussion
This data set results from the first effort to obtain miRNA expression profiling data of external ear development in mammals. This data should prove useful to understand the role of miRNAs in normal external ear development in mammals and might hint over miRNA involvement in the etiology of microtia [4] Data was validated by Whole mount in situ hybridization in a subset of miRNAs whose mRNA targets have been associated with external ear development and present clear differential spatiotemporal expression patterns (mmu-miR-10a, mmu-miR-200c and mmu-miR-205) [1], [2], [3], [5]. The microarray data presented in this work has been deposited in Gene Expression Omnibus through GEO Series with accession number GSE64945.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
Financial support: Federal Funding from the Instituto Nacional de Pediatría 053/2013. U. Juarez received a CONACyT scholarship 270077 from the Posgrado en Ciencias Biológicas, UNAM. We would like to thank the personnel of the animal care facility at the Instituto Nacional de Pediatría, for their professional work with the mice and to Dr. Benilde García de Teresa for her valuable comments and revision of the manuscript.
References
- 1.Garzon R., Pichiorri F., Palumbo T., Iuliano R., Cimmino A., Aqeilan R., Volinia S., Bhatt D., Alder H., Marcucci G., Calin G.A., Liu C.G., Bloomfield C.D., Andreeff M., Croce C.M. MicroRNA fingerprints during human megakaryocytopoiesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavalas A., Stunder M., Lumsden A., Rijli F.M., Krumlauf R., Chambon P. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development. 1998;125:1123–1136. doi: 10.1242/dev.125.6.1123. [DOI] [PubMed] [Google Scholar]
- 3.Li C., Hao S., Wang H., Jin L., Qing F., Zheng F., Zhang P., Chen L., Ma D., Zhang T. MicroRNA expression profiling and target genes study in congenital microtia. Int. J. Pediatr. Otorhinolaryngol. 2013;77:483–487. doi: 10.1016/j.ijporl.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Luquetti D.V., Heike C.L., Hing A.V., Cunningham M.L., Cox T.C. Microtia: epidemiology and genetics. Am. J. Med. Genet. 2012;158A:24–39. doi: 10.1002/ajmg.a.34352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres L., Juárez U., García L., Miranda-Ríos J., Frias S. External ear microRNA expression profiles during mouse development. Int. J. Dev. Biol. 2015;59:497–503. doi: 10.1387/ijdb.150124sf. [DOI] [PubMed] [Google Scholar]