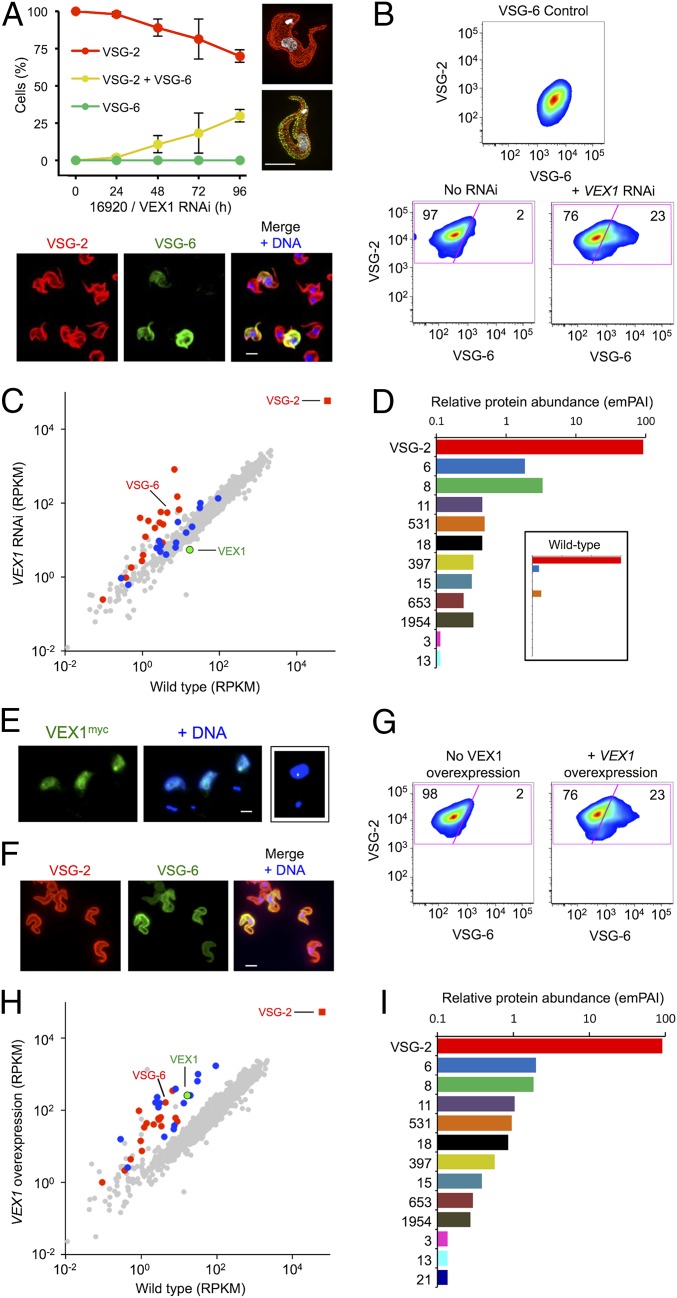
Fig. 3.
VEX1 controls VSG allelic exclusion in bloodstream-form cells. (A) Immunofluorescence microscopy analysis of VSG expression. Cells were stained with α-VSG-2 and α-VSG-6 and counted daily during VEX1-RNAi. The 3D-SIM images show a wild-type control cell and a cell expressing both VSGs. The images below the plot show cells following VEX1-RNAi (72 h). (Scale bars, 5 μm.) (B) Flow-cytometry analysis of VSG expression following VEX1-RNAi (72 h). Numbers indicate percentage of cells in each quadrant. VSG-6 expressers serve as a control. n = 10,000 cells in each case. (C) RNA-seq analysis following VEX1-RNAi (72 h). Values are averages for a pair of independent strains (Dataset S1). Red circles, silent VSGs; red square, active VSG; blue circles, procyclins and procyclin-associated genes. RPKM, reads per kilobase of transcript per million mapped reads. (D) Quantitative mass spectrometry analysis of surface VSGs following VEX1-RNAi (72 h) (SI Appendix, Table S2). (Inset) Wild-type cells for comparison. emPAI, exponentially modified Protein Abundance Index. (E) Immunofluorescence microscopy of overexpressed and ectopic VEX1myc (72 h). (Scale bar, 2 μm.) (Right) Sequestered VEX1 for comparison. (F) Immunofluorescence microscopy analysis of VSG expression following VEX1 overexpression (72 h). (Scale bar, 5 μm.) (G) Flow cytometry following VEX1 overexpression (72 h). Other details are as in B. (H) RNA-seq analysis following VEX1 overexpression (72 h). Other details are as in C. (I) Quantitative mass spectrometry analysis of surface VSGs following VEX1 overexpression (72 h) (SI Appendix, Table S2).