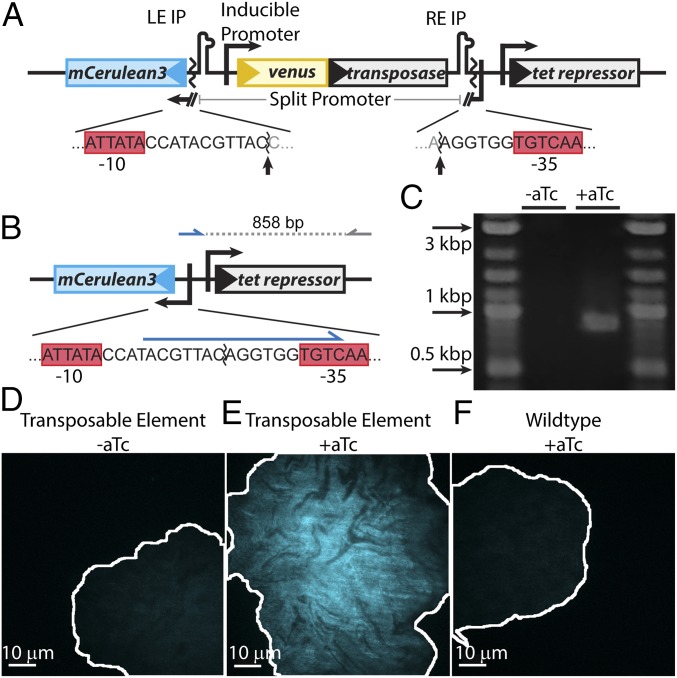
Fig. 1.
Design and validation of the TE system. (A) The promoter for mCerulean3 is interrupted by the transposable element, the ends of which are demarcated by left end and right end imperfect palindromic sequences (LE IP and RE IP). The transposase, tnpA (gray), is expressed from the promoter PLtetO1, which is inducible with aTc. The sequences of the promoter/TE junction and −10 and −35 sequences (red boxes) are shown below the diagram, and the sites cleaved by transposase are indicated by arrows. (B) Upon excision, the promoter for mCerulean3 is reconstituted and the cell fluoresces blue. The sequence of the reconstituted promoter is shown below the diagram. A primer designed to bind to the unique sequence formed after promoter reconstitution (blue arrow) was used to verify excision by PCR, generating an 858-bp amplicon. (C) PCR amplification using these primers only generates the 858-bp product upon induction, thus verifying excision. (D–F) Colony morphology after growth on agarose pads. Uninduced TE-carrying cells (D) and wild-type cells exposed to 20 ng/mL aTc (F) show homogeneous, low blue autofluorescence. Conversely, TE-carrying cells induced with 20 ng/mL aTc (E) show bright, inhomogeneous blue fluorescence. The brightness scale for all three images is identical. The borders of the colonies are outlined in white.