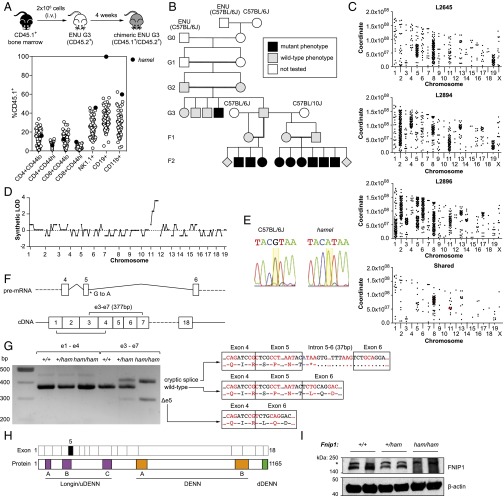
Fig. 1.
Recessive B-cell deficiency associated with a splice donor variant in Fnip1. (A) Schematic of the competitive reconstitution screen and identification of the hamel phenotype. (B) Initial generations of the hamel pedigree, including mapping outcrosses. (C) Single-nucleotide variant distribution across the genomes of three affected mice and those shared by all three. Mutations affecting protein-coding sense are highlighted in red. (D) Simulated LOD score generated by chromosomal mapping. (E) Capillary-sequencing trace of a splice donor variant in Fnip1 (yellow highlight). (F) Schematic of the Fnip1 transcript (ENSMUST00000046835) and the location of the hamel mutation and positions of amplicons generated in G. (G) Fnip1 BM cDNA PCR amplification and sequencing, demonstrating the presence of two major alternate splice products in Fnip1 homozygous mutants. (H) Domain structure of the FNIP1 protein, with corresponding coding exons displayed above. Exon 5 (skipped in one alternate splice product) is highlighted in black fill. DENN, differentially expressed in neoplastic versus normal cells. (I) anti-FNIP1 Western blot on splenocyte lysates of the indicated genotypes. Asterisk indicates a nonspecific band.