The African trypanosome, Trypanosoma brucei, is a causative agent of African Trypanosomiasis (also known as “sleeping sickness” in humans and “nagana” in cattle) and imposes an enormous economic burden in regions of Sub-Saharan Africa. T. brucei is transmitted to the mammalian host through the bite of the tsetse fly. In the mammal, it survives extracellularly in the bloodstream, eventually migrating into the central nervous system and causing coma and death. To survive in the bloodstream of its mammalian host, T. brucei must evade the host immune system. The parasite is covered by a dense, variant surface glycoprotein (VSG) coat that shields other epitopes on the cell surface from antibodies produced by the host immune system (reviewed in refs. 1 and 2). A strong antibody response is mounted against the VSG upon entry of the parasite into the bloodstream. However, the parasite harbors thousands of variants of the VSG gene and periodically switches the particular variant that is expressed in a process called antigenic variation (known colloquially in T. brucei as “switching”) (1).
VSG genes are transcribed from one of ∼15 telomeric bloodstream expression sites (BESs), only one of which is transcriptionally active at any given time. Transcription of VSG genes occurs within a discrete nuclear structure, called the expression site body (ESB) and is driven by Pol I (1). Switching from expression of one VSG to a new VSG can occur by multiple mechanisms, including transcriptional activation of a new BES and recombinatorial mechanisms (1). Analysis of T. brucei populations in mouse models of infection has revealed that a population of trypanosomes can contain parasites expressing as many as 66 different VSGs at any given time (3). However, although the repertoire of VSGs expressed in a population of trypanosomes is quite diverse, a single trypanosome expresses just one VSG on its surface at any given time, except when actively undergoing a switch. Monoallelic expression of a single gene variant is found across diverse biological systems, including allelic exclusion of Ig genes, olfactory receptor expression, rhodopsin gene use in the retina, and X- chromosome inactivation. Thus, understanding the regulation of monoallelic expression in T. brucei can lend insight into how these regulatory processes evolved.
The PNAS paper, “VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes” by Glover et al. (4), seeks to understand the problem of how one VSG gene is kept transcriptionally active while the rest are silenced. This problem has long been an area of active inquiry, and many proteins have been identified that play a role in maintaining monoallelic expression of VSG genes. In particular, as in other biological systems (5, 6), chromatin structure plays a role. BESs are long polycistronic units that contain a number of expression site-associated genes (ESAGs) upstream of the VSG gene, which is found ∼45 kb downstream from the promoter (2). The active BES is depleted of nucleosomes, whereas inactive BESs are comparatively nucleosome-rich (7). Depletion of histone H3, H1, or H3.V causes increased transcription at inactive BES promoters (8, 9) or at VSGs themselves (10, 11). Histone deposition factors and nucleosome remodelers have also been shown to be important for maintaining silencing at these sites (9, 12). Interfering with proteins that read, write, or erase modifications on the histone tails can disrupt silencing of telomeric reporter genes or inactive BESs, depending on the factor (9, 13–17). Other factors that have been shown to be important for maintaining silencing of inactive BESs include those involved in telomere maintenance and DNA replication (18, 19). However, with the notable exception of the histone methyltransferase, Dot1B, and the telomeric factor, Rap1, perturbation of these factors increases transcription at inactive BESs but does not lead to expression of two different VSG proteins on the surface of a single parasite.
Glover et al. (4) attacked this problem in an unbiased fashion by screening an RNAi library for factors that, when depleted, produced increased transcription of a telomeric reporter gene. This is a particularly elegant approach given that a large majority of genes in the trypanosome genome have uncharacterized functions. Glover et al.’s efforts led to the identification of a gene, Tb927.11.16920, which they named “VEX1” for VSG EXclusion 1. At the protein level, depletion of VEX1 by RNAi caused expression of two different VSGs at levels comparable to those previously reported for Dot1B deletion (16). To expand their analysis, the authors (4) carried out RNA-seq experiments showing that a large number of inactive BES VSG genes and metacyclic expression site VSGs were transcribed at much higher levels in VEX1-depleted cells when compared with controls. Impressively, the authors generated proteomic data to verify that increased transcription of VSGs at these inactive sites resulted in proteins that were translated and transported to the cell surface, something that has never been demonstrated for this number of VSGs to date. Although several groups have reported increased transcription of VSGs at inactive sites, Glover et al. are the first group to report translation and transport to the surface for a large number of inactive VSGs following depletion of a single factor.
A central question raised by this paper (4) and others is whether the loss of silencing seen at the population level reflects loss of silencing of all of the inactive loci in a single cell, or whether the lower levels of RNA transcript from inactive VSGs measured at the population level reflect only one or a few loci becoming stochastically derepressed in each cell. If the derepressed locus is chosen stochastically, the transcript level for the derepressed VSG might be quite high in the individual cell, but low when measured at the population level, provided each cell is derepressing one or a few loci randomly. The fact that an inactive VSG has a protein abundance that is roughly two orders-of-magnitude lower than the active VSG at the population level, yet appears to be expressed highly when measured by flow cytometry, may favor this latter model, but because antibody affinities can be variable, this can’t be definitively concluded. On the other hand, one would expect only ∼6% of cells to express the inactive VSG if only one BES was derepressed in each cell and BESs were chosen randomly. Instead, Glover et al. (4) find that 23% of the cells express the inactive VSG-6 following depletion of VEX1; this might argue for multiple BESs getting derepressed simultaneously or, instead, a hierarchy or preference for a particular BES becoming derepressed as a result of nuclear positioning, and so forth. An important next step will be to perform single-cell RNA-seq experiments to ascertain whether all loci are equally derepressed in every cell, or whether one or a subset of loci become derepressed in each individual cell. It remains to be seen whether more than two VSGs can be expressed on the surface of the same cell following depletion of VEX1 or any other factor. If multiple VSG proteins are produced in a single cell, but only two are found on the surface, this might indicate
Although the molecular mechanism of action of VEX1 with other proteins or RNA remains to be established, Glover et al. have made an important step forward in identifying a main player in the maintenance of monoallelic expression in T. brucei.
that a second mechanism operates posttranscriptionally to ensure that a single cell does not express a diverse repertoire of VSGs at the surface. For example, additional regulatory mechanisms may be in play during trafficking of VSGs. Can different VSGs be packed together on the surface? Do certain VSGs pack better or less well with others? If this is the case, physical constraints may prevent more than two VSGs from being expressed on a single parasite cell surface.
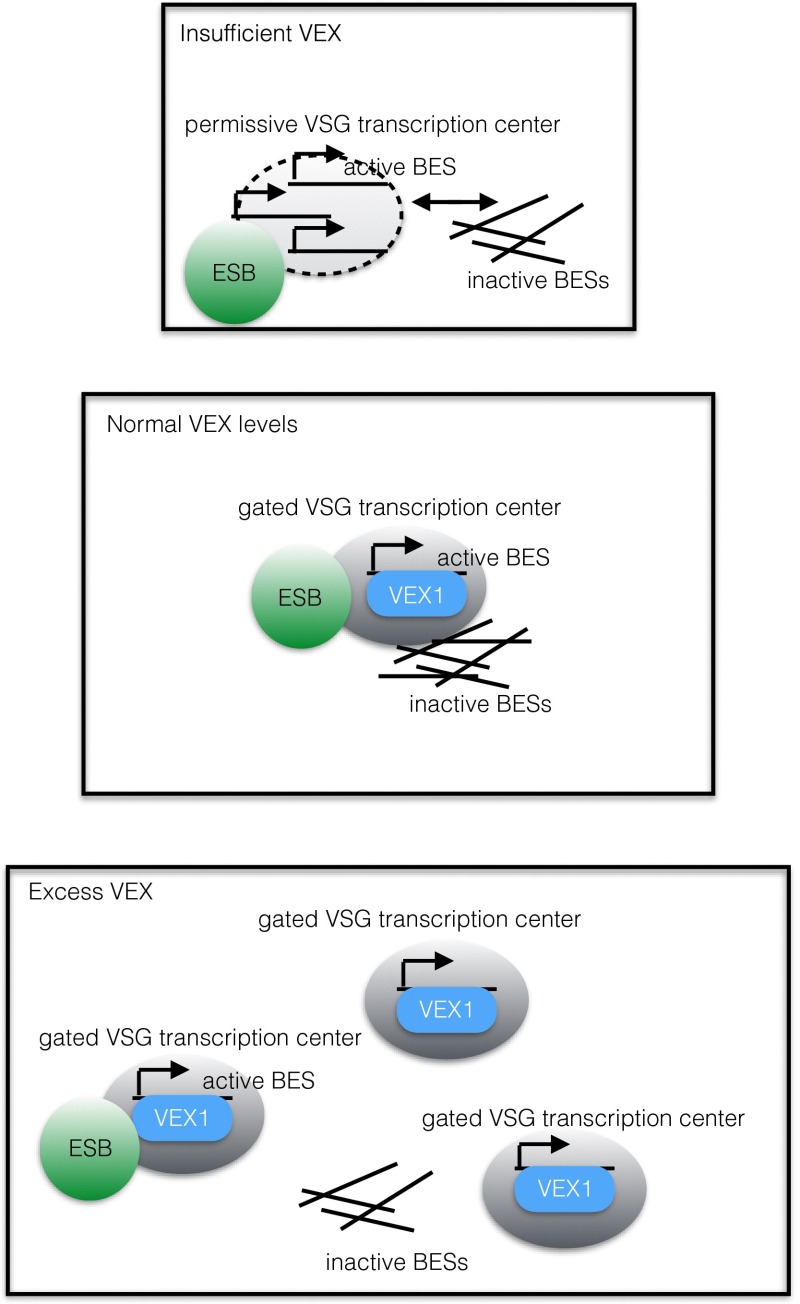
Glover et al. (4) demonstrate that the VEX1 protein exists in a single focus that overlaps the telomeric protein, TRF2, and coincides with—but does not overlap—the ESB in bloodstream cells. In contrast, the protein is diffused throughout the nucleus in insect-stage cells, when VSG is no longer expressed. Seemingly incongruously, the loss of VEX1 leads to loss of monoallelic expression, but overexpression of VEX1 also results in this phenotype, leading the authors to postulate that VEX1 has a dual role as a positive driver of VSG transcription and a factor that inhibits transcription of VSG genes at inactive sites. The key to this apparent paradox is an observation by the authors that in clones that stably express two VSGs, two VEX1 foci appear in a higher fraction of the cells, but one is often distal to the ESB. Thus, VEX1 seems capable of establishing gated “centers” of VSG transcription.
If one imagines VEX1 as a gatekeeper for entry to an ESB-associated VSG transcription “center,” then loss of VEX1 would allow a “free-for-all” entry of additional BESs within the ESB-associated center and access to the transcription factors housed within it [Pol I, class I transcription factor A (CITFA), or nucleoplasmin-like protein (NLP) (12, 20), and possibly others], resulting in increased transcription of VSGs at the newly associated BESs. Conversely, overabundance of VEX1 would establish additional gated centers for VSG expression (which might not necessarily coincide with the ESB). Although the outcomes might be similar, the mechanisms that drive loss of monoallelic expression during VEX1 depletion vs. forced multiallelic expression during VEX1 overexpression are qualitatively different. In this model, the wild-type situation would involve one active BES within a VEX1-gated, ESB-associated VSG-transcribing center, with VEX1 being the limiting factor for establishing additional centers (Fig. 1).
Fig. 1.
A hypothetical model for VEX regulation. Each square represents the nucleus of a cell. Three situations are depicted (normal VEX1 expression, depletion of VEX1, or overexpression of VEX1). Green circle: ESB; blue oval: VEX-1 VSG transcribing center. See text for a more detailed description.
VEX1’s only characterized protein motif is a SWIM domain, making its mechanism of action a particularly thorny problem. Using an artificial telomere system, Glover et al. (4) observed that placement of two genes with the same UTR near the telomere results in transcriptional silencing of the telomere proximal gene, whereas this is not the case if the two genes harbor different UTRs. Silencing of the telomere proximal gene is disrupted when VEX1 is depleted. Based on these results, the authors propose a “winner-takes-all” model wherein VEX1 is sequestered, establishing a single, active BES. Silencing of VSGs is then mediated via homology within VSG UTRs and telomeric sequences. Although this is an intriguing model, the telomeric bicistronic reporter system used in these experiments is limited in that it contains an rDNA promoter rather than a VSG BES promoter that is placed closer to the VSG than the one in a BES, no ESAG sequences are present, and the UTRs used are not VSG UTRs. Experiments using reporter genes within inactive BESs might help to elucidate whether VEX1 is directly or indirectly maintaining silencing via homologous BES transcripts or DNA sequences. One could imagine a scenario like that seen in X-chromosome inactivation, where long-noncoding RNAs emanating from BES sequences regulate monoallelic expression. Although the molecular mechanism of action of VEX1 with other proteins or RNA remains to be established, Glover et al. have made an important step forward in identifying a main player in the maintenance of monoallelic expression in T. brucei. Their work has opened up an exciting new area for these authors and others to understand how this intriguing protein regulates antigenic variation.
Footnotes
The authors declare no conflict of interest.
See companion article on page 7225.
References
- 1.Hovel-Miner G, Mugnier M, Papavasiliou FN, Pinger J, Schulz D. A host-pathogen interaction reduced to first principles: Antigenic variation in T. brucei. Results Probl Cell Differ. 2015;57:23–46. doi: 10.1007/978-3-319-20819-0_2. [DOI] [PubMed] [Google Scholar]
- 2.Horn D. Antigenic variation in African trypanosomes. Mol Biochem Parasitol. 2014;195(2):123–129. doi: 10.1016/j.molbiopara.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mugnier MR, Cross GAM, Papavasiliou FN. The in vivo dynamics of antigenic variation in Trypanosoma brucei. Science. 2015;347(6229):1470–1473. doi: 10.1126/science.aaa4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glover L, Hutchinson S, Alsford S, Horn D. VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc Natl Acad Sci USA. 2016;113:7225–7230. doi: 10.1073/pnas.1600344113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlissel MS, Stanhope-Baker P. Accessibility and the developmental regulation of V(D)J recombination. Semin Immunol. 1997;9(3):161–170. doi: 10.1006/smim.1997.0066. [DOI] [PubMed] [Google Scholar]
- 6.Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145(4):555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueiredo LM, Cross GAM. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot Cell. 2010;9(1):148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Povelones ML, Gluenz E, Dembek M, Gull K, Rudenko G. Histone H1 plays a role in heterochromatin formation and VSG expression site silencing in Trypanosoma brucei. PLoS Pathog. 2012;8(11):e1003010. doi: 10.1371/journal.ppat.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alsford S, Horn D. Cell-cycle-regulated control of VSG expression site silencing by histones and histone chaperones ASF1A and CAF-1b in Trypanosoma brucei. Nucleic Acids Res. 2012;40(20):10150–10160. doi: 10.1093/nar/gks813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulz D, Zaringhalam M, Papavasiliou FN, Kim H-S. Base J and H3.V regulate transcriptional termination in Trypanosoma brucei. PLoS Genet. 2016;12(1):e1005762. doi: 10.1371/journal.pgen.1005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds D, et al. Histone H3 variant regulates RNA polymerase II transcription termination and dual strand transcription of siRNA loci in Trypanosoma brucei. PLoS Genet. 2016;12(1):e1005758. doi: 10.1371/journal.pgen.1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narayanan MS, et al. NLP is a novel transcription regulator involved in VSG expression site control in Trypanosoma brucei. Nucleic Acids Res. 2011;39(6):2018–2031. doi: 10.1093/nar/gkq950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alsford S, Kawahara T, Isamah C, Horn D. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol Microbiol. 2007;63(3):724–736. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q-P, Kawahara T, Horn D. Histone deacetylases play distinct roles in telomeric VSG expression site silencing in African trypanosomes. Mol Microbiol. 2010;77(5):1237–1245. doi: 10.1111/j.1365-2958.2010.07284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawahara T, et al. Two essential MYST-family proteins display distinct roles in histone H4K10 acetylation and telomeric silencing in trypanosomes. Mol Microbiol. 2008;69(4):1054–1068. doi: 10.1111/j.1365-2958.2008.06346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueiredo LM, Janzen CJ, Cross GAM. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6(7):e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz D, et al. Bromodomain proteins contribute to maintenance of bloodstream form stage identity in the African trypanosome. PLoS Biol. 2015;13(12):e1002316–e1002338. doi: 10.1371/journal.pbio.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H-S, Park SH, Günzl A, Cross GAM. MCM-BP is required for repression of life-cycle specific genes transcribed by RNA polymerase I in the mammalian infectious form of Trypanosoma brucei. PLoS One. 2013;8(2):e57001. doi: 10.1371/journal.pone.0057001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137(1):99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen TN, Nguyen BN, Lee JH, Panigrahi AK, Günzl A. Characterization of a novel class I transcription factor A (CITFA) subunit that is indispensable for transcription by the multifunctional RNA polymerase I of Trypanosoma brucei. Eukaryot Cell. 2012;11(12):1573–1581. doi: 10.1128/EC.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]